Gene
KWMTBOMO07165
Pre Gene Modal
BGIBMGA010556
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_1-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.979
Sequence
CDS
ATGTTTCGGCGGAAATCCTGCTACATATTTCAAGATGATAAACTTATTGAAACGTTGACTATTAAGGAATCACTTACGATAGCAGCGGAACTGAAACTTGGAAATCATATCAGTGCGAGTCAAAAACGACAACGCGTAGAACAAATAATAATGTCTCTTGGGCTGTACGCAGTGGAGAATACTAAAGCCGGGAACCTATCAGGCGGACAAAAAAAAAGACTAACAATAGGTCTGGAACTAGTCAGCGATCCGCCCGTTATGTTCCTCGATGAACCAACAAGTGGACTAGATATTTCGGTAGCAAAAAAATTAGTCAATTTACTTCACCTGTTAGCACGGCAAGGACGAACGATGGTGATAACTATGCATCAACCTTCACTCAGTCTACTTCAGATGGTGGACAGGCTATACGCAGTCGTTGCAGGAAAGTGTGCTTATATGGGCTCCGTGCCTCTGCTACTGCCATATTTAAACCAATTGAACCTACCAGACTTAAGGTGCTTGTCGTATCATGACCCTGTTGAATATTTAATAGAAATCAGTGGGACCTATGAAGAAAAATTAGTAGAAAAGTCTAAAAATGGAAGTAACAATTGTTGGGTTACTGAAGCGACTGATAGTCCAATTCTAGAAAGACCGAAACTACTTAGAATTACTTCAACAAATGAGGAAATTAGTTTAACACTATTAACAGCTCCCAAAGAGAATTCGACAAATAAAATTATTTTGGCCCTGAAAAGCACATACTCTACCTCATTTTGGAAACAATTCATCACATTGACAAGAAGATCTGTATTAAGTACGTGGCGAGAACCTTCATTTACGTTAACATTCATAAGTATTCACGGATTCATGGCATTGTTTATTGGCATCCTATTCAAGAATGTTGGCGAAGACGTCGAAAAAGTCAGAGATAACTACAACTTTCTGTACTTCAGTCTGATGTTCCTTATGTTCAACGCATTTAGTGCCGTCTCAATCAGATTTCCGGAACAAATCCCCGTTACCCGAAGAGAACATTTCAACCGTTGGTATACGACAGGCGCTTTCTACGCGTCAACTTTACTCTCTACGCTACCCATACAGACCGTGTGCACCCTTCTATACTCAATTCTCGTATATTGGATGACAGGACAATCTCCGAATTTAATTCGATTTACAATATTCAATTTCACTTTGCTGTTAGTCTCATACGTATCACTATGTATGGGTCTCCTTAATGGATCAATATTTAACGTAAAGAATGGAGTTATATTTGGTCCGCTAATGATTATGCCGTTCACTATATTCTCCGGGTTCTTCTTGCGTTACCGCGATGCGCCGTATTTCTTCCGCTGGATGTTTCAAATATCATTCTTGAAACACGGACTCGTTGGACTCGTGCTCTCCGTCTTCGGTATGGGCCGGTCTAACTTCAGCTGCCCAAAAATATTCTGTTACTACACTTACCCTACGCAGTTATTAATAGACAACCACATGGACAAAGAAGAATTTACATTAGCGATTGGAGCACTTTTCGCTATCAGCTTCGCATTGATTGCGACTTCTTTCGTGATCCTCAAAATTAGACTGAAGAGTAAATGGTGA
Protein
MFRRKSCYIFQDDKLIETLTIKESLTIAAELKLGNHISASQKRQRVEQIIMSLGLYAVENTKAGNLSGGQKKRLTIGLELVSDPPVMFLDEPTSGLDISVAKKLVNLLHLLARQGRTMVITMHQPSLSLLQMVDRLYAVVAGKCAYMGSVPLLLPYLNQLNLPDLRCLSYHDPVEYLIEISGTYEEKLVEKSKNGSNNCWVTEATDSPILERPKLLRITSTNEEISLTLLTAPKENSTNKIILALKSTYSTSFWKQFITLTRRSVLSTWREPSFTLTFISIHGFMALFIGILFKNVGEDVEKVRDNYNFLYFSLMFLMFNAFSAVSIRFPEQIPVTRREHFNRWYTTGAFYASTLLSTLPIQTVCTLLYSILVYWMTGQSPNLIRFTIFNFTLLLVSYVSLCMGLLNGSIFNVKNGVIFGPLMIMPFTIFSGFFLRYRDAPYFFRWMFQISFLKHGLVGLVLSVFGMGRSNFSCPKIFCYYTYPTQLLIDNHMDKEEFTLAIGALFAISFALIATSFVILKIRLKSKW
Summary
Uniprot
A0A194R0A1
A0A2A4K0D4
A0A3S2P675
A0A194PI62
A0A2H1V3X0
A0A1E1WQ41
+ More
K7IMI8 A0A1L8E4V1 A0A2M4BIK8 D2A233 A0A2M4BHN6 A0A182MW61 A0A2M4BGS2 A0A2M4BGY9 A0A182S4M3 A0A2A3ET89 A0A0U9HU30 A0A088A0P1 A0A182KBU9 A0A2M4BEZ2 A0A0C9RBC3 A0A182XFM6 A0A1S4H158 A0A2M4AF23 A0A1Y9GLI2 A0A182Q8Q3 E2BE50 A0A1S4H221 A0A1S4H1Z0 A0A1S4H226 A0A232FLJ7 A0A182VHC8 A0A2M4AMD6 A0A182JLA7 A0A0L7QX62 A0A0N0BL17 A0A182GYV8 V5GU74 B0X144 A0A084VPQ0 Q16Y88 W5J4Y2 A0A026WG29 A0A1Q3FJ30 A0A1Q3FIP9 A0A087ZKJ5 A0A2M4CZ66 U5EUW9 A0A182YMF1 A0A1W4X6U4 U4UDC0 N6TU97 A0A1W4XHI2 A0A154NZ61 A0A3R5SLE9 A0A2P8Y9C3 A0A182VUV2 A0A151WW34 E2A2I6 A0A3Q0J6W2 A0A3Q0J1V8 A0A1Y1NBY8 U5ETU1 A0A182P7E7 A0A3L8DF84 A0A0K8U8N1 F4WD70 A0A146KSC3 A0A158P165 A0A1Y1KDU5 A0A023F9W8 D2A232 A0A1B0GJ00 V5GHI9 E0W387 A0A2J7RCU0 A0A067QV98 A0A146M2T0 A0A2J7PQ37 A0A067QKG8 A0A0A9Y4U0 A0A0C9S320 E2A2I4 A0A151WWP2 E9IZD6 D1ZZE5 E0W3T6 K7IQL0 E9IZD3 A0A0C9RP34 A0A158P0V0 A0A1J1IRZ3 A0A2Z5UB45 H9J6F2 A0A1B6CQQ0 A0A151JZF0 A0A151IVA2 A0A195AUJ8 A0A1S4EBY9
K7IMI8 A0A1L8E4V1 A0A2M4BIK8 D2A233 A0A2M4BHN6 A0A182MW61 A0A2M4BGS2 A0A2M4BGY9 A0A182S4M3 A0A2A3ET89 A0A0U9HU30 A0A088A0P1 A0A182KBU9 A0A2M4BEZ2 A0A0C9RBC3 A0A182XFM6 A0A1S4H158 A0A2M4AF23 A0A1Y9GLI2 A0A182Q8Q3 E2BE50 A0A1S4H221 A0A1S4H1Z0 A0A1S4H226 A0A232FLJ7 A0A182VHC8 A0A2M4AMD6 A0A182JLA7 A0A0L7QX62 A0A0N0BL17 A0A182GYV8 V5GU74 B0X144 A0A084VPQ0 Q16Y88 W5J4Y2 A0A026WG29 A0A1Q3FJ30 A0A1Q3FIP9 A0A087ZKJ5 A0A2M4CZ66 U5EUW9 A0A182YMF1 A0A1W4X6U4 U4UDC0 N6TU97 A0A1W4XHI2 A0A154NZ61 A0A3R5SLE9 A0A2P8Y9C3 A0A182VUV2 A0A151WW34 E2A2I6 A0A3Q0J6W2 A0A3Q0J1V8 A0A1Y1NBY8 U5ETU1 A0A182P7E7 A0A3L8DF84 A0A0K8U8N1 F4WD70 A0A146KSC3 A0A158P165 A0A1Y1KDU5 A0A023F9W8 D2A232 A0A1B0GJ00 V5GHI9 E0W387 A0A2J7RCU0 A0A067QV98 A0A146M2T0 A0A2J7PQ37 A0A067QKG8 A0A0A9Y4U0 A0A0C9S320 E2A2I4 A0A151WWP2 E9IZD6 D1ZZE5 E0W3T6 K7IQL0 E9IZD3 A0A0C9RP34 A0A158P0V0 A0A1J1IRZ3 A0A2Z5UB45 H9J6F2 A0A1B6CQQ0 A0A151JZF0 A0A151IVA2 A0A195AUJ8 A0A1S4EBY9
Pubmed
EMBL
KQ460883
KPJ11218.1
NWSH01000345
PCG77233.1
RSAL01000299
RVE42784.1
+ More
KQ459603 KPI92738.1 ODYU01000563 SOQ35538.1 GDQN01002103 JAT88951.1 GFDF01000522 JAV13562.1 GGFJ01003749 MBW52890.1 KQ971338 EFA01493.1 GGFJ01003411 MBW52552.1 AXCM01001117 GGFJ01003109 MBW52250.1 GGFJ01003122 MBW52263.1 KZ288191 PBC34512.1 GARF01000012 JAC88925.1 GGFJ01002473 MBW51614.1 GBYB01004176 JAG73943.1 AAAB01008839 GGFK01006001 MBW39322.1 APCN01004583 AXCN02000853 GL447755 EFN86037.1 NNAY01000050 OXU31535.1 GGFK01008457 MBW41778.1 KQ414705 KOC63207.1 KQ435687 KOX81384.1 JXUM01021309 JXUM01021310 KQ560597 KXJ81700.1 GALX01000747 JAB67719.1 DS232252 EDS38493.1 ATLV01015022 KE524999 KFB39944.1 CH477522 EAT39580.1 ADMH02002133 ETN58463.1 KK107235 EZA54898.1 GFDL01007466 JAV27579.1 GFDL01007606 JAV27439.1 GGFL01006446 MBW70624.1 GANO01002142 JAB57729.1 KB632081 ERL88591.1 APGK01052119 APGK01052120 APGK01052121 APGK01052122 APGK01052123 APGK01052124 KB741209 ENN72850.1 KQ434783 KZC04862.1 MH172510 QAA95917.1 PYGN01000782 PSN40866.1 KQ982691 KYQ52093.1 GL436038 EFN72389.1 GEZM01007026 JAV95472.1 GANO01002631 JAB57240.1 QOIP01000009 RLU19117.1 GDHF01029292 GDHF01014089 JAI23022.1 JAI38225.1 GL888084 EGI67878.1 GDHC01019236 JAP99392.1 ADTU01005828 GEZM01087692 JAV58531.1 GBBI01000426 JAC18286.1 EFA02729.1 AJWK01017948 AJWK01017949 AJWK01017950 GALX01004992 JAB63474.1 DS235882 EEB20093.1 NEVH01005883 PNF38654.1 KK853336 KDR08337.1 GDHC01004531 GDHC01001750 JAQ14098.1 JAQ16879.1 NEVH01022640 PNF18457.1 KK853235 KDR09575.1 GBHO01016416 GBHO01016415 GBHO01016413 GBHO01016411 GBRD01006668 GBRD01006666 JAG27188.1 JAG27189.1 JAG27191.1 JAG27193.1 JAG59153.1 GBYB01015296 JAG85063.1 EFN72387.1 KYQ52091.1 GL767121 EFZ14046.1 EFA01863.2 DS235883 EEB20292.1 EFZ14056.1 GBYB01015295 GBYB01015297 JAG85062.1 JAG85064.1 ADTU01005818 ADTU01005819 ADTU01005820 CVRI01000057 CRL02482.1 LC322265 BBB02488.1 BABH01019639 BABH01019640 GEDC01021537 JAS15761.1 KQ981414 KYN41883.1 KQ980916 KYN11448.1 KQ976738 KYM75695.1
KQ459603 KPI92738.1 ODYU01000563 SOQ35538.1 GDQN01002103 JAT88951.1 GFDF01000522 JAV13562.1 GGFJ01003749 MBW52890.1 KQ971338 EFA01493.1 GGFJ01003411 MBW52552.1 AXCM01001117 GGFJ01003109 MBW52250.1 GGFJ01003122 MBW52263.1 KZ288191 PBC34512.1 GARF01000012 JAC88925.1 GGFJ01002473 MBW51614.1 GBYB01004176 JAG73943.1 AAAB01008839 GGFK01006001 MBW39322.1 APCN01004583 AXCN02000853 GL447755 EFN86037.1 NNAY01000050 OXU31535.1 GGFK01008457 MBW41778.1 KQ414705 KOC63207.1 KQ435687 KOX81384.1 JXUM01021309 JXUM01021310 KQ560597 KXJ81700.1 GALX01000747 JAB67719.1 DS232252 EDS38493.1 ATLV01015022 KE524999 KFB39944.1 CH477522 EAT39580.1 ADMH02002133 ETN58463.1 KK107235 EZA54898.1 GFDL01007466 JAV27579.1 GFDL01007606 JAV27439.1 GGFL01006446 MBW70624.1 GANO01002142 JAB57729.1 KB632081 ERL88591.1 APGK01052119 APGK01052120 APGK01052121 APGK01052122 APGK01052123 APGK01052124 KB741209 ENN72850.1 KQ434783 KZC04862.1 MH172510 QAA95917.1 PYGN01000782 PSN40866.1 KQ982691 KYQ52093.1 GL436038 EFN72389.1 GEZM01007026 JAV95472.1 GANO01002631 JAB57240.1 QOIP01000009 RLU19117.1 GDHF01029292 GDHF01014089 JAI23022.1 JAI38225.1 GL888084 EGI67878.1 GDHC01019236 JAP99392.1 ADTU01005828 GEZM01087692 JAV58531.1 GBBI01000426 JAC18286.1 EFA02729.1 AJWK01017948 AJWK01017949 AJWK01017950 GALX01004992 JAB63474.1 DS235882 EEB20093.1 NEVH01005883 PNF38654.1 KK853336 KDR08337.1 GDHC01004531 GDHC01001750 JAQ14098.1 JAQ16879.1 NEVH01022640 PNF18457.1 KK853235 KDR09575.1 GBHO01016416 GBHO01016415 GBHO01016413 GBHO01016411 GBRD01006668 GBRD01006666 JAG27188.1 JAG27189.1 JAG27191.1 JAG27193.1 JAG59153.1 GBYB01015296 JAG85063.1 EFN72387.1 KYQ52091.1 GL767121 EFZ14046.1 EFA01863.2 DS235883 EEB20292.1 EFZ14056.1 GBYB01015295 GBYB01015297 JAG85062.1 JAG85064.1 ADTU01005818 ADTU01005819 ADTU01005820 CVRI01000057 CRL02482.1 LC322265 BBB02488.1 BABH01019639 BABH01019640 GEDC01021537 JAS15761.1 KQ981414 KYN41883.1 KQ980916 KYN11448.1 KQ976738 KYM75695.1
Proteomes
UP000053240
UP000218220
UP000283053
UP000053268
UP000002358
UP000007266
+ More
UP000075883 UP000075900 UP000242457 UP000005203 UP000075881 UP000076407 UP000075840 UP000075886 UP000008237 UP000215335 UP000075903 UP000075880 UP000053825 UP000053105 UP000069940 UP000249989 UP000002320 UP000030765 UP000008820 UP000000673 UP000053097 UP000076408 UP000192223 UP000030742 UP000019118 UP000076502 UP000245037 UP000075920 UP000075809 UP000000311 UP000079169 UP000075885 UP000279307 UP000007755 UP000005205 UP000092461 UP000009046 UP000235965 UP000027135 UP000183832 UP000005204 UP000078541 UP000078492 UP000078540
UP000075883 UP000075900 UP000242457 UP000005203 UP000075881 UP000076407 UP000075840 UP000075886 UP000008237 UP000215335 UP000075903 UP000075880 UP000053825 UP000053105 UP000069940 UP000249989 UP000002320 UP000030765 UP000008820 UP000000673 UP000053097 UP000076408 UP000192223 UP000030742 UP000019118 UP000076502 UP000245037 UP000075920 UP000075809 UP000000311 UP000079169 UP000075885 UP000279307 UP000007755 UP000005205 UP000092461 UP000009046 UP000235965 UP000027135 UP000183832 UP000005204 UP000078541 UP000078492 UP000078540
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A194R0A1
A0A2A4K0D4
A0A3S2P675
A0A194PI62
A0A2H1V3X0
A0A1E1WQ41
+ More
K7IMI8 A0A1L8E4V1 A0A2M4BIK8 D2A233 A0A2M4BHN6 A0A182MW61 A0A2M4BGS2 A0A2M4BGY9 A0A182S4M3 A0A2A3ET89 A0A0U9HU30 A0A088A0P1 A0A182KBU9 A0A2M4BEZ2 A0A0C9RBC3 A0A182XFM6 A0A1S4H158 A0A2M4AF23 A0A1Y9GLI2 A0A182Q8Q3 E2BE50 A0A1S4H221 A0A1S4H1Z0 A0A1S4H226 A0A232FLJ7 A0A182VHC8 A0A2M4AMD6 A0A182JLA7 A0A0L7QX62 A0A0N0BL17 A0A182GYV8 V5GU74 B0X144 A0A084VPQ0 Q16Y88 W5J4Y2 A0A026WG29 A0A1Q3FJ30 A0A1Q3FIP9 A0A087ZKJ5 A0A2M4CZ66 U5EUW9 A0A182YMF1 A0A1W4X6U4 U4UDC0 N6TU97 A0A1W4XHI2 A0A154NZ61 A0A3R5SLE9 A0A2P8Y9C3 A0A182VUV2 A0A151WW34 E2A2I6 A0A3Q0J6W2 A0A3Q0J1V8 A0A1Y1NBY8 U5ETU1 A0A182P7E7 A0A3L8DF84 A0A0K8U8N1 F4WD70 A0A146KSC3 A0A158P165 A0A1Y1KDU5 A0A023F9W8 D2A232 A0A1B0GJ00 V5GHI9 E0W387 A0A2J7RCU0 A0A067QV98 A0A146M2T0 A0A2J7PQ37 A0A067QKG8 A0A0A9Y4U0 A0A0C9S320 E2A2I4 A0A151WWP2 E9IZD6 D1ZZE5 E0W3T6 K7IQL0 E9IZD3 A0A0C9RP34 A0A158P0V0 A0A1J1IRZ3 A0A2Z5UB45 H9J6F2 A0A1B6CQQ0 A0A151JZF0 A0A151IVA2 A0A195AUJ8 A0A1S4EBY9
K7IMI8 A0A1L8E4V1 A0A2M4BIK8 D2A233 A0A2M4BHN6 A0A182MW61 A0A2M4BGS2 A0A2M4BGY9 A0A182S4M3 A0A2A3ET89 A0A0U9HU30 A0A088A0P1 A0A182KBU9 A0A2M4BEZ2 A0A0C9RBC3 A0A182XFM6 A0A1S4H158 A0A2M4AF23 A0A1Y9GLI2 A0A182Q8Q3 E2BE50 A0A1S4H221 A0A1S4H1Z0 A0A1S4H226 A0A232FLJ7 A0A182VHC8 A0A2M4AMD6 A0A182JLA7 A0A0L7QX62 A0A0N0BL17 A0A182GYV8 V5GU74 B0X144 A0A084VPQ0 Q16Y88 W5J4Y2 A0A026WG29 A0A1Q3FJ30 A0A1Q3FIP9 A0A087ZKJ5 A0A2M4CZ66 U5EUW9 A0A182YMF1 A0A1W4X6U4 U4UDC0 N6TU97 A0A1W4XHI2 A0A154NZ61 A0A3R5SLE9 A0A2P8Y9C3 A0A182VUV2 A0A151WW34 E2A2I6 A0A3Q0J6W2 A0A3Q0J1V8 A0A1Y1NBY8 U5ETU1 A0A182P7E7 A0A3L8DF84 A0A0K8U8N1 F4WD70 A0A146KSC3 A0A158P165 A0A1Y1KDU5 A0A023F9W8 D2A232 A0A1B0GJ00 V5GHI9 E0W387 A0A2J7RCU0 A0A067QV98 A0A146M2T0 A0A2J7PQ37 A0A067QKG8 A0A0A9Y4U0 A0A0C9S320 E2A2I4 A0A151WWP2 E9IZD6 D1ZZE5 E0W3T6 K7IQL0 E9IZD3 A0A0C9RP34 A0A158P0V0 A0A1J1IRZ3 A0A2Z5UB45 H9J6F2 A0A1B6CQQ0 A0A151JZF0 A0A151IVA2 A0A195AUJ8 A0A1S4EBY9
PDB
6FFC
E-value=2.24728e-47,
Score=478
Ontologies
GO
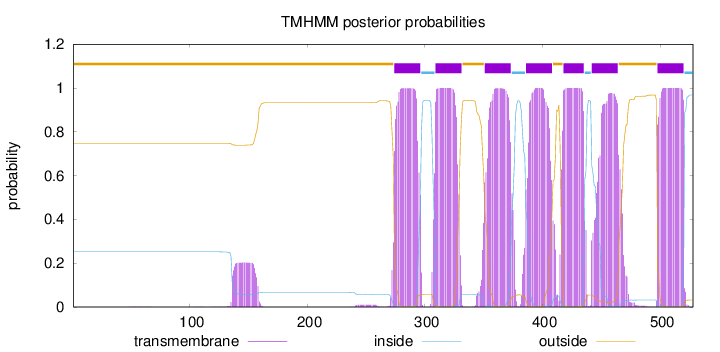
Topology
Length:
528
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
158.05786
Exp number, first 60 AAs:
0.00226
Total prob of N-in:
0.25237
outside
1 - 273
TMhelix
274 - 296
inside
297 - 308
TMhelix
309 - 331
outside
332 - 350
TMhelix
351 - 373
inside
374 - 385
TMhelix
386 - 408
outside
409 - 417
TMhelix
418 - 435
inside
436 - 441
TMhelix
442 - 464
outside
465 - 497
TMhelix
498 - 520
inside
521 - 528
Population Genetic Test Statistics
Pi
196.510925
Theta
160.402783
Tajima's D
-0.975648
CLR
118.795115
CSRT
0.142592870356482
Interpretation
Uncertain
