Gene
KWMTBOMO07164
Pre Gene Modal
BGIBMGA010555
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.972
Sequence
CDS
ATGACGCCCGGCCTCGCCGAAGATATTGGGAACGGCAATAACAATGTCGTGCCGCTGCTGCCAAGAACAACGGTGGAATTGGAGTTCAGAAACATCAAGTGTACTATTAATACATACAGTCTCATTAAATTTAAAATGGAACAAAGACAAATCCTGAAGGGAGTGTCAGGATGCTTCAAGCCAGGAGCGCTAACTGCCATCATGGGTCCGTCGGGTGCGGGCAAGACAACGCTCTTGAACATACTCGCCGGGTATTCGACGCAAGGGGCTAAAGGTGAGATAGCCGTGAATGGCGCGTGTACCTGCGCCTCGCAGTCAAGCGGACGCGGCGGTGCACGATACATCCGCCAGCACGATGACCTGCGTGCGTATCTGACGGTCTACGAAGCAATGTCTCTCGCAGCTGCTCTTAAACTGGCCGACTTCTCAAGACATGAAAGAAAGGAAAAGGTTTATGAAATTCTAGCTCTTTTGGGTTTAAGCGCTGCGGCGGAAACACTATGCCGTGACCTGTCCGGAGGCCAAAAGAGAAGATTGGCTGTAGCTTTAGAATTGCTCTCAGATCCATCCCTCATGTTCCTGGATGAGCCTACTACCGGCTTGGACTCATCTGCATCGGCGTCATTGATAACCTTACTGAACAATCTCGCTCGAGGAGGTCGTACACTAATAGCTACAATTCATCAACCTTCTGCCCTCATATTCGAGAAGATCGACAATGTGTACTGCCTGGAGGCTGGTAGGACTCTGTACAATGGGCCCAGAGCAGCCCTTGTTCCGGCACTCGCCAGGGCCGGTCTACAGTGTCCGTCGTATCATAATCCGGCCGATTTTCTAATGGAAGTTGCGTCCGGTGAATACGACGTAGACCTTATAAAGGTGGCAAAGAAGATGAGTCAATTCAAAACAGACGACGCTGTCAATGGCAGCTGGGTAGAGATTCCAACCATACCAGCACTGCCTTCTTCTTTAGATATAAGTACTGAAATGAAGTCGAAACATTTCGGAGAGAGTCCCAAAATGCGTAAACTCTCAAAGAACATATACAAGCCATCAGGGCAGGTCCGGCAGACTGCCATTTTGCTGTATAGAAACTATTTGATTACTACAAGAAATTATGCTCTATTCATGTACCGAGTTGCTGCTCACGTTGTTATCGCTTTAATATTTGGATATCTTTACTCCGGAGTCGGCGGCGAAGCTAGTTCAGTACTAGGGAACTATGTATATCTGTATGGATCGATGTTGCTAACCGTGTACACTGGAAAAATGTCCGTTACATTATCATTTCCCTTAGAAATGGAAATACTCAAAGGCGAATACTTCAATCGATGGTATAACCTGGCGCCGTATATATTTTCCATATTAGCAGTGGAACTACCATTTCAAATGATATCCTGCGTGTCCTATGTCGTTCTATCGTATTGGTTGACCGACAACCCGTTGGAGCCGACGCGAGCAACGTTGTTCTTGGCCACAATCGTAGCCACGTCTTTATGCGCACAAGCCTGGGGATATTTCATCGGTTCGACAACACCCACAAAGATCGCAGTGTTCATAGGACCAGTGCTGGCGTGTCTGTTTTCAGTATTCGGATTCTGTCTCGCACTATCTGACACGCCACGTTACTTCCGCTGGCTCCACTACATCTCGTACTTCAGAGCCGGCTTTCATGTCGCTGTATACTCCGTTTACGGTTTTAATAGAAAAAAACTTCAATGCTCTAAACTATATTGTCAATTCGTAACGCCAAAGACCTTCTTAAAACAAATGGACATGGAAGAGGTGGACATCGGTGGAAACATGACTATAGTTATATTAACTACAGTGTTAATGCATTTATTAACGTGCGCTGCTCTTTGGTACAGACTTAATAAACGATGA
Protein
MTPGLAEDIGNGNNNVVPLLPRTTVELEFRNIKCTINTYSLIKFKMEQRQILKGVSGCFKPGALTAIMGPSGAGKTTLLNILAGYSTQGAKGEIAVNGACTCASQSSGRGGARYIRQHDDLRAYLTVYEAMSLAAALKLADFSRHERKEKVYEILALLGLSAAAETLCRDLSGGQKRRLAVALELLSDPSLMFLDEPTTGLDSSASASLITLLNNLARGGRTLIATIHQPSALIFEKIDNVYCLEAGRTLYNGPRAALVPALARAGLQCPSYHNPADFLMEVASGEYDVDLIKVAKKMSQFKTDDAVNGSWVEIPTIPALPSSLDISTEMKSKHFGESPKMRKLSKNIYKPSGQVRQTAILLYRNYLITTRNYALFMYRVAAHVVIALIFGYLYSGVGGEASSVLGNYVYLYGSMLLTVYTGKMSVTLSFPLEMEILKGEYFNRWYNLAPYIFSILAVELPFQMISCVSYVVLSYWLTDNPLEPTRATLFLATIVATSLCAQAWGYFIGSTTPTKIAVFIGPVLACLFSVFGFCLALSDTPRYFRWLHYISYFRAGFHVAVYSVYGFNRKKLQCSKLYCQFVTPKTFLKQMDMEEVDIGGNMTIVILTTVLMHLLTCAALWYRLNKR
Summary
Uniprot
A0A2I7ZPD4
H9JM03
A0A2A4JZM7
A0A212ERD4
A0A194R129
A0A194PHE9
+ More
A0A0F6PQM6 J3JYT7 A0A0C9PTI5 D1ZZD4 K7IMI9 A0A158P0V1 A0A1L8E493 A0A2A3ES35 A0A182QIU7 A0A0M9AE00 A0A1S4H177 A0A1Y9GLD0 A0A182RGB6 A0A182L5H8 A0A182VBS3 A0A2M4A4S3 A0A2M4BG18 A0A2M3Z427 A0A182F591 A0A151WWQ8 A0A3L8DGX7 A0A084VPQ5 F4WD67 A0A0U9HSG3 A0A1B0GJ01 A0A182VUV1 A0A0J7NSR5 A0A195AU49 A0A067QSH6 A0A2M4CZ66 A0A087ZKJ5 A0A2J7RCT9 A0A3R5WTC2 A0A182P7E9 A0A195CXU5 A0A1Q3F9H8 A0A1Q3F9M4 B0X143 W5J4Y2 J9K8S0 A0A182G5I5 A0A0U9HVF4 A0A2H8TWH9 A0A232FLB2 A0A161UF76 A0A0L7QXE2 B4KKU3 E9IZD7 A0A151JAR2 A0A154P153 J9JYQ2 A0A0A9WA49 A0A146M7D3 A0A2R7W3D0 N6TMY7 A0A224X9J0 A0A023F2V9 A0A1B6LW75 E9HL04 E0W3T6 A0A1S4N1I2 J9K4C5 D2A233 A0A1W4YJT2 A0A2D0ST94 V5GU74 A0A151J3S1 A0A195FTJ6 A0A2S2Q1D6 A0A154PC74 A0A3Q2TSE0 A0A1S4EBY9 M3ZUQ1 A0A3Q2Q0I9 E0W387 A0A151WQM4 A0A3Q3MKV8 A0A2I4D7R2 A0A1Y1NBY8 A0A3Q3IU81 A0A3R5SMP8 A0A146XMQ4 A0A226ESE3 A0A2J7RCU4 A0A3B1KJ86 A0A3P9K5H7 A0A0A7ARN0 A0A0A9W0J1 A0A3B4DS14 A0A3Q1JAB2
A0A0F6PQM6 J3JYT7 A0A0C9PTI5 D1ZZD4 K7IMI9 A0A158P0V1 A0A1L8E493 A0A2A3ES35 A0A182QIU7 A0A0M9AE00 A0A1S4H177 A0A1Y9GLD0 A0A182RGB6 A0A182L5H8 A0A182VBS3 A0A2M4A4S3 A0A2M4BG18 A0A2M3Z427 A0A182F591 A0A151WWQ8 A0A3L8DGX7 A0A084VPQ5 F4WD67 A0A0U9HSG3 A0A1B0GJ01 A0A182VUV1 A0A0J7NSR5 A0A195AU49 A0A067QSH6 A0A2M4CZ66 A0A087ZKJ5 A0A2J7RCT9 A0A3R5WTC2 A0A182P7E9 A0A195CXU5 A0A1Q3F9H8 A0A1Q3F9M4 B0X143 W5J4Y2 J9K8S0 A0A182G5I5 A0A0U9HVF4 A0A2H8TWH9 A0A232FLB2 A0A161UF76 A0A0L7QXE2 B4KKU3 E9IZD7 A0A151JAR2 A0A154P153 J9JYQ2 A0A0A9WA49 A0A146M7D3 A0A2R7W3D0 N6TMY7 A0A224X9J0 A0A023F2V9 A0A1B6LW75 E9HL04 E0W3T6 A0A1S4N1I2 J9K4C5 D2A233 A0A1W4YJT2 A0A2D0ST94 V5GU74 A0A151J3S1 A0A195FTJ6 A0A2S2Q1D6 A0A154PC74 A0A3Q2TSE0 A0A1S4EBY9 M3ZUQ1 A0A3Q2Q0I9 E0W387 A0A151WQM4 A0A3Q3MKV8 A0A2I4D7R2 A0A1Y1NBY8 A0A3Q3IU81 A0A3R5SMP8 A0A146XMQ4 A0A226ESE3 A0A2J7RCU4 A0A3B1KJ86 A0A3P9K5H7 A0A0A7ARN0 A0A0A9W0J1 A0A3B4DS14 A0A3Q1JAB2
Pubmed
EMBL
KY775299
AUS94651.1
BABH01005636
BABH01005637
BABH01005638
NWSH01000345
+ More
PCG77226.1 AGBW02013032 OWR44063.1 KQ460883 KPJ11219.1 KQ459603 KPI92737.1 KM244041 AKC42334.1 BT128418 AEE63375.1 GBYB01004673 JAG74440.1 KQ971338 EFA01865.1 ADTU01005831 ADTU01005832 ADTU01005833 ADTU01005834 GFDF01000516 JAV13568.1 KZ288191 PBC34510.1 AXCN02000853 KQ435687 KOX81383.1 AAAB01008839 APCN01004583 GGFK01002409 MBW35730.1 GGFJ01002859 MBW52000.1 GGFM01002519 MBW23270.1 KQ982691 KYQ52095.1 QOIP01000009 RLU19118.1 ATLV01015024 KE524999 KFB39949.1 GL888084 EGI67875.1 GARF01000020 JAC88919.1 AJWK01017950 AJWK01017951 AJWK01017952 AJWK01017953 LBMM01001964 KMQ95470.1 KQ976738 KYM75701.1 KK853336 KDR08338.1 GGFL01006446 MBW70624.1 NEVH01005883 PNF38651.1 MH172511 QAA95918.1 KQ977141 KYN05402.1 GFDL01010825 JAV24220.1 GFDL01010775 JAV24270.1 DS232252 EDS38492.1 ADMH02002133 ETN58463.1 ABLF02039577 JXUM01043676 JXUM01043677 JXUM01043678 JXUM01043679 KQ561371 KXJ78817.1 GARF01000063 JAC88894.1 GFXV01006464 MBW18269.1 NNAY01000050 OXU31534.1 KF828799 QKKF02019844 AIN44116.1 RZF39315.1 KQ414705 KOC63209.1 CH933807 EDW11673.2 GL767121 EFZ14064.1 KQ979216 KYN22241.1 KQ434783 KZC04860.1 ABLF02039565 GBHO01038276 GBHO01038272 GBHO01038271 GBHO01018987 GBHO01018983 GBHO01018980 GBRD01017575 GBRD01007511 JAG05328.1 JAG05332.1 JAG05333.1 JAG24617.1 JAG24621.1 JAG24624.1 JAG48252.1 GDHC01002965 JAQ15664.1 KK854233 PTY13490.1 APGK01028919 APGK01028920 APGK01028921 APGK01028922 APGK01028923 APGK01028924 KB740713 ENN79393.1 GFTR01007441 JAW08985.1 GBBI01003271 JAC15441.1 GEBQ01012041 JAT27936.1 GL732674 EFX67564.1 DS235883 EEB20292.1 AAZO01007341 ABLF02036674 EFA01493.1 GALX01000747 JAB67719.1 KQ980249 KYN17092.1 KQ981276 KYN43612.1 GGMS01002375 MBY71578.1 KQ434864 KZC09024.1 DS235882 EEB20093.1 KQ982821 KYQ50166.1 GEZM01007026 JAV95472.1 MH172516 QAA95923.1 GCES01042983 JAR43340.1 LNIX01000002 OXA59711.1 PNF38648.1 KF906302 AHK05667.1 GBHO01041657 JAG01947.1
PCG77226.1 AGBW02013032 OWR44063.1 KQ460883 KPJ11219.1 KQ459603 KPI92737.1 KM244041 AKC42334.1 BT128418 AEE63375.1 GBYB01004673 JAG74440.1 KQ971338 EFA01865.1 ADTU01005831 ADTU01005832 ADTU01005833 ADTU01005834 GFDF01000516 JAV13568.1 KZ288191 PBC34510.1 AXCN02000853 KQ435687 KOX81383.1 AAAB01008839 APCN01004583 GGFK01002409 MBW35730.1 GGFJ01002859 MBW52000.1 GGFM01002519 MBW23270.1 KQ982691 KYQ52095.1 QOIP01000009 RLU19118.1 ATLV01015024 KE524999 KFB39949.1 GL888084 EGI67875.1 GARF01000020 JAC88919.1 AJWK01017950 AJWK01017951 AJWK01017952 AJWK01017953 LBMM01001964 KMQ95470.1 KQ976738 KYM75701.1 KK853336 KDR08338.1 GGFL01006446 MBW70624.1 NEVH01005883 PNF38651.1 MH172511 QAA95918.1 KQ977141 KYN05402.1 GFDL01010825 JAV24220.1 GFDL01010775 JAV24270.1 DS232252 EDS38492.1 ADMH02002133 ETN58463.1 ABLF02039577 JXUM01043676 JXUM01043677 JXUM01043678 JXUM01043679 KQ561371 KXJ78817.1 GARF01000063 JAC88894.1 GFXV01006464 MBW18269.1 NNAY01000050 OXU31534.1 KF828799 QKKF02019844 AIN44116.1 RZF39315.1 KQ414705 KOC63209.1 CH933807 EDW11673.2 GL767121 EFZ14064.1 KQ979216 KYN22241.1 KQ434783 KZC04860.1 ABLF02039565 GBHO01038276 GBHO01038272 GBHO01038271 GBHO01018987 GBHO01018983 GBHO01018980 GBRD01017575 GBRD01007511 JAG05328.1 JAG05332.1 JAG05333.1 JAG24617.1 JAG24621.1 JAG24624.1 JAG48252.1 GDHC01002965 JAQ15664.1 KK854233 PTY13490.1 APGK01028919 APGK01028920 APGK01028921 APGK01028922 APGK01028923 APGK01028924 KB740713 ENN79393.1 GFTR01007441 JAW08985.1 GBBI01003271 JAC15441.1 GEBQ01012041 JAT27936.1 GL732674 EFX67564.1 DS235883 EEB20292.1 AAZO01007341 ABLF02036674 EFA01493.1 GALX01000747 JAB67719.1 KQ980249 KYN17092.1 KQ981276 KYN43612.1 GGMS01002375 MBY71578.1 KQ434864 KZC09024.1 DS235882 EEB20093.1 KQ982821 KYQ50166.1 GEZM01007026 JAV95472.1 MH172516 QAA95923.1 GCES01042983 JAR43340.1 LNIX01000002 OXA59711.1 PNF38648.1 KF906302 AHK05667.1 GBHO01041657 JAG01947.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000002358 UP000005205 UP000242457 UP000075886 UP000053105 UP000075840 UP000075900 UP000075882 UP000075903 UP000069272 UP000075809 UP000279307 UP000030765 UP000007755 UP000092461 UP000075920 UP000036403 UP000078540 UP000027135 UP000235965 UP000075885 UP000078542 UP000002320 UP000000673 UP000007819 UP000069940 UP000249989 UP000215335 UP000291343 UP000053825 UP000009192 UP000078492 UP000076502 UP000019118 UP000000305 UP000009046 UP000192224 UP000221080 UP000078541 UP000265000 UP000079169 UP000002852 UP000261640 UP000192220 UP000261600 UP000198287 UP000018467 UP000265180 UP000261440 UP000265040
UP000002358 UP000005205 UP000242457 UP000075886 UP000053105 UP000075840 UP000075900 UP000075882 UP000075903 UP000069272 UP000075809 UP000279307 UP000030765 UP000007755 UP000092461 UP000075920 UP000036403 UP000078540 UP000027135 UP000235965 UP000075885 UP000078542 UP000002320 UP000000673 UP000007819 UP000069940 UP000249989 UP000215335 UP000291343 UP000053825 UP000009192 UP000078492 UP000076502 UP000019118 UP000000305 UP000009046 UP000192224 UP000221080 UP000078541 UP000265000 UP000079169 UP000002852 UP000261640 UP000192220 UP000261600 UP000198287 UP000018467 UP000265180 UP000261440 UP000265040
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2I7ZPD4
H9JM03
A0A2A4JZM7
A0A212ERD4
A0A194R129
A0A194PHE9
+ More
A0A0F6PQM6 J3JYT7 A0A0C9PTI5 D1ZZD4 K7IMI9 A0A158P0V1 A0A1L8E493 A0A2A3ES35 A0A182QIU7 A0A0M9AE00 A0A1S4H177 A0A1Y9GLD0 A0A182RGB6 A0A182L5H8 A0A182VBS3 A0A2M4A4S3 A0A2M4BG18 A0A2M3Z427 A0A182F591 A0A151WWQ8 A0A3L8DGX7 A0A084VPQ5 F4WD67 A0A0U9HSG3 A0A1B0GJ01 A0A182VUV1 A0A0J7NSR5 A0A195AU49 A0A067QSH6 A0A2M4CZ66 A0A087ZKJ5 A0A2J7RCT9 A0A3R5WTC2 A0A182P7E9 A0A195CXU5 A0A1Q3F9H8 A0A1Q3F9M4 B0X143 W5J4Y2 J9K8S0 A0A182G5I5 A0A0U9HVF4 A0A2H8TWH9 A0A232FLB2 A0A161UF76 A0A0L7QXE2 B4KKU3 E9IZD7 A0A151JAR2 A0A154P153 J9JYQ2 A0A0A9WA49 A0A146M7D3 A0A2R7W3D0 N6TMY7 A0A224X9J0 A0A023F2V9 A0A1B6LW75 E9HL04 E0W3T6 A0A1S4N1I2 J9K4C5 D2A233 A0A1W4YJT2 A0A2D0ST94 V5GU74 A0A151J3S1 A0A195FTJ6 A0A2S2Q1D6 A0A154PC74 A0A3Q2TSE0 A0A1S4EBY9 M3ZUQ1 A0A3Q2Q0I9 E0W387 A0A151WQM4 A0A3Q3MKV8 A0A2I4D7R2 A0A1Y1NBY8 A0A3Q3IU81 A0A3R5SMP8 A0A146XMQ4 A0A226ESE3 A0A2J7RCU4 A0A3B1KJ86 A0A3P9K5H7 A0A0A7ARN0 A0A0A9W0J1 A0A3B4DS14 A0A3Q1JAB2
A0A0F6PQM6 J3JYT7 A0A0C9PTI5 D1ZZD4 K7IMI9 A0A158P0V1 A0A1L8E493 A0A2A3ES35 A0A182QIU7 A0A0M9AE00 A0A1S4H177 A0A1Y9GLD0 A0A182RGB6 A0A182L5H8 A0A182VBS3 A0A2M4A4S3 A0A2M4BG18 A0A2M3Z427 A0A182F591 A0A151WWQ8 A0A3L8DGX7 A0A084VPQ5 F4WD67 A0A0U9HSG3 A0A1B0GJ01 A0A182VUV1 A0A0J7NSR5 A0A195AU49 A0A067QSH6 A0A2M4CZ66 A0A087ZKJ5 A0A2J7RCT9 A0A3R5WTC2 A0A182P7E9 A0A195CXU5 A0A1Q3F9H8 A0A1Q3F9M4 B0X143 W5J4Y2 J9K8S0 A0A182G5I5 A0A0U9HVF4 A0A2H8TWH9 A0A232FLB2 A0A161UF76 A0A0L7QXE2 B4KKU3 E9IZD7 A0A151JAR2 A0A154P153 J9JYQ2 A0A0A9WA49 A0A146M7D3 A0A2R7W3D0 N6TMY7 A0A224X9J0 A0A023F2V9 A0A1B6LW75 E9HL04 E0W3T6 A0A1S4N1I2 J9K4C5 D2A233 A0A1W4YJT2 A0A2D0ST94 V5GU74 A0A151J3S1 A0A195FTJ6 A0A2S2Q1D6 A0A154PC74 A0A3Q2TSE0 A0A1S4EBY9 M3ZUQ1 A0A3Q2Q0I9 E0W387 A0A151WQM4 A0A3Q3MKV8 A0A2I4D7R2 A0A1Y1NBY8 A0A3Q3IU81 A0A3R5SMP8 A0A146XMQ4 A0A226ESE3 A0A2J7RCU4 A0A3B1KJ86 A0A3P9K5H7 A0A0A7ARN0 A0A0A9W0J1 A0A3B4DS14 A0A3Q1JAB2
PDB
6FFC
E-value=1.32149e-52,
Score=524
Ontologies
GO
Topology
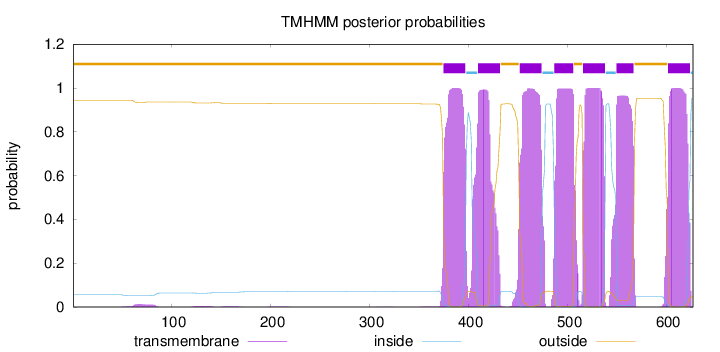
Length:
627
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
151.73981
Exp number, first 60 AAs:
0.02282
Total prob of N-in:
0.05547
outside
1 - 374
TMhelix
375 - 397
inside
398 - 409
TMhelix
410 - 432
outside
433 - 451
TMhelix
452 - 474
inside
475 - 486
TMhelix
487 - 506
outside
507 - 515
TMhelix
516 - 538
inside
539 - 549
TMhelix
550 - 567
outside
568 - 601
TMhelix
602 - 624
inside
625 - 627
Population Genetic Test Statistics
Pi
184.973288
Theta
153.823421
Tajima's D
0.173752
CLR
0.428448
CSRT
0.420378981050947
Interpretation
Uncertain
