Gene
KWMTBOMO07162
Pre Gene Modal
BGIBMGA010554
Annotation
PREDICTED:_neural_cell_adhesion_molecule_1-like_isoform_X2_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.235 PlasmaMembrane Reliability : 1.247
Sequence
CDS
ATGTTTGGACCACCAAACCTTTTGTCAACGGAGGTGGACGACGATGAGCCATTTCTTGAAATATTGGCACGATCCACGATTTACAATACTGGAGAGAGAAAGGCGATTTTTTGCAGAGGAAAAAATTTAGTAGAGCCAATTAAATGGTACTCTCCTTCGGGAAAACTAATAGAAGCGCAATCTTTGAAAAACAAGAGAGTATACGTTGAAAGAATTGTTGAGGAATCAGGGGAACATGCGCCGTTAATCATCCAATACATCAAGATTGCGGACAACGGTAACTGGACTTGCAGGTCTGGAGATTTGAGTGAAACCATACAATTCATTGTCGGAGAAAAGGTGAACATTACTAACAGGAATGTTGAAAGAGAAGGGGAAGAAGGAAAATCAATTGTGCTGACCTGTGAAGCCAAGGGACATCCGACACCGGTCGTAATTTGGTACAAAGAAAATTCTCGAATCCCTAATGATACTAAAAAGTACGAGCACAGGGCTGACAACTCGTTGGAGATCAAGAATCTGAACCACGCTGACGTCGGACTGTACGTGTGCAAAGTTAGACAAAAGGCGCTGTCCTACTACACAGACAAAACAGTCCGTCTTTCTGTCCTACATAAGCCGATTCTGTATAACAACTTCACGGGAACGATCGACACCACGAAATACAAGACTGAAGAAGTTTACGCAATTATGAATGAAACGAAGAACATCACATGCAGCGCTATCGCAAACCCCCCGCCGACTTACACCTGGTACAGAAGAAACAATGGTTACGACGATGATGTTCCTATCACTGACGAGGAGATGGTGGTCACTTCAGAAAACGGTACATATTCAGTACTCATTTTGAGGATGCGCGATAACAGTTCAATCGGCGAGTACAAATGCACAGCGAACAACATCAAGGGGAAAGAATCTATTATATTCCATGTAAATCCAGCAGCTAAACCCAACAAACCTGATTTTGTAAGCTTTCTTTACGCTAACACTACGACTCTGACTTTCAATGTTTCCTGCTCGAGCTGCCCAATGGAACTAGAAGAAGATAGGTCACCCGACCCAGCAAACCTTACTGTTAAAGGATATTCATTCCAAATGGTTCCTGCCCAAGAAAATTACCCTCCAGATTGGGATTCTGCTCAAGATTTCGAAGTGGACATCACAGATCCTAACGACACGTTGTTCACGGTCGGCCCACTAGCCAACAAGACAAAGTACCACGTCCGCATCCGAAGCTGCAACGCGGCCGGCTACTCTGAGTGGGTGTACTTCGAGACACTACTAGAGACGAGCCACGCTGACAGGATCATCGCAGCGATCGCTGTCATCTTAGCGGCCTTCACGATTACAAGCTATTTCTAA
Protein
MFGPPNLLSTEVDDDEPFLEILARSTIYNTGERKAIFCRGKNLVEPIKWYSPSGKLIEAQSLKNKRVYVERIVEESGEHAPLIIQYIKIADNGNWTCRSGDLSETIQFIVGEKVNITNRNVEREGEEGKSIVLTCEAKGHPTPVVIWYKENSRIPNDTKKYEHRADNSLEIKNLNHADVGLYVCKVRQKALSYYTDKTVRLSVLHKPILYNNFTGTIDTTKYKTEEVYAIMNETKNITCSAIANPPPTYTWYRRNNGYDDDVPITDEEMVVTSENGTYSVLILRMRDNSSIGEYKCTANNIKGKESIIFHVNPAAKPNKPDFVSFLYANTTTLTFNVSCSSCPMELEEDRSPDPANLTVKGYSFQMVPAQENYPPDWDSAQDFEVDITDPNDTLFTVGPLANKTKYHVRIRSCNAAGYSEWVYFETLLETSHADRIIAAIAVILAAFTITSYF
Summary
Uniprot
H9JM02
A0A2A4JGX1
A0A2H1V173
A0A194PJ37
A0A212F9U5
A0A194R0J4
+ More
A0A3S2N6K1 A0A182NEE1 A0A182J736 A0A084VPE7 A0A0D3QDB5 A0A182RGG1 A0A182QG23 A0A182MRE5 A0A0D3QCN6 A0A0D3QCX8 A0A0D3QDB1 A0A0D3QCY8 A0A0D3QCP3 A0A0D3QCM8 A0A0D3QDA7 A0A0D3QDI4 A0A0D3QDJ3 A0A0D3QCY3 Q7QG58 A0A182VGZ9 A0A182F5W4 A0A182X7N4 A0A182L5N5 A0A182VUZ9 A0A182IE33 A0A182H555 A0A194R5S7 A0A182UAU5 A0A182K6S0 A0A182P383 B0W5V2 A0A1S4J637 C3YY39 A0A194PNN6 A0A1W4UIW6 B4KLE0 D2A2M5 A0A182H630 A0A3B0JX29 B3MTZ8 A0A336LT49 B4I3C2 B4Q324 B4NZ06 A0A0J9TFK6 Q9VR25 B4JQQ7 B3N495 Q29LA6 A0A0M4E6D9 B4GQC8 A0A146M1R4 W5JJ33 A0A087Z3M8 A0A212F9T9 A0A1Y1N5J0 A0A0L0CMK1 A0A1B0F9W5 A0A146LR12 A0A0L0C706 B4LUV2 A0A0M3QSZ8 A0A1A9YQX8 A0A1B0AZB3 B4MTW1 W8AST7 W8BD55 B4NW33 A0A3B0JV17 A0A3B0JGE0 A0A3B0K2T3 A0A2H1VBZ7 A0A0Q9XIZ7 B4KIY3 A0A1I8MMA3 A0A0A1XD89 A0A2A4JG26 B4IDE7 B4MVI2 B3N901 B4LVH9 A0A0K8W9X7 B4Q7L2 A0A0J9QU46 Q59E14 D0Z750 B4JAH4 A0A1I8M2C2 Q29LN2 W8ALP2 B3MJD9 A0A2J7R8N1
A0A3S2N6K1 A0A182NEE1 A0A182J736 A0A084VPE7 A0A0D3QDB5 A0A182RGG1 A0A182QG23 A0A182MRE5 A0A0D3QCN6 A0A0D3QCX8 A0A0D3QDB1 A0A0D3QCY8 A0A0D3QCP3 A0A0D3QCM8 A0A0D3QDA7 A0A0D3QDI4 A0A0D3QDJ3 A0A0D3QCY3 Q7QG58 A0A182VGZ9 A0A182F5W4 A0A182X7N4 A0A182L5N5 A0A182VUZ9 A0A182IE33 A0A182H555 A0A194R5S7 A0A182UAU5 A0A182K6S0 A0A182P383 B0W5V2 A0A1S4J637 C3YY39 A0A194PNN6 A0A1W4UIW6 B4KLE0 D2A2M5 A0A182H630 A0A3B0JX29 B3MTZ8 A0A336LT49 B4I3C2 B4Q324 B4NZ06 A0A0J9TFK6 Q9VR25 B4JQQ7 B3N495 Q29LA6 A0A0M4E6D9 B4GQC8 A0A146M1R4 W5JJ33 A0A087Z3M8 A0A212F9T9 A0A1Y1N5J0 A0A0L0CMK1 A0A1B0F9W5 A0A146LR12 A0A0L0C706 B4LUV2 A0A0M3QSZ8 A0A1A9YQX8 A0A1B0AZB3 B4MTW1 W8AST7 W8BD55 B4NW33 A0A3B0JV17 A0A3B0JGE0 A0A3B0K2T3 A0A2H1VBZ7 A0A0Q9XIZ7 B4KIY3 A0A1I8MMA3 A0A0A1XD89 A0A2A4JG26 B4IDE7 B4MVI2 B3N901 B4LVH9 A0A0K8W9X7 B4Q7L2 A0A0J9QU46 Q59E14 D0Z750 B4JAH4 A0A1I8M2C2 Q29LN2 W8ALP2 B3MJD9 A0A2J7R8N1
Pubmed
19121390
26354079
22118469
24438588
25552603
12364791
+ More
20966253 26483478 18563158 17994087 18362917 19820115 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 26823975 20920257 28004739 26108605 24495485 25315136 25830018
20966253 26483478 18563158 17994087 18362917 19820115 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 15632085 26823975 20920257 28004739 26108605 24495485 25315136 25830018
EMBL
BABH01005632
NWSH01001507
PCG71026.1
ODYU01000209
SOQ34600.1
KQ459603
+ More
KPI92734.1 AGBW02009558 OWR50507.1 KQ460883 KPJ11222.1 RSAL01000299 RVE42787.1 ATLV01015001 KE524999 KFB39841.1 KP274524 AJC98670.1 AXCN02000859 AXCM01000195 KP274530 AJC98676.1 KP274521 AJC98667.1 KP274519 AJC98665.1 KP274531 AJC98677.1 KP274532 AJC98678.1 KP274513 KP274515 KP274516 KP274517 KP274523 KP274527 KP274529 AJC98659.1 AJC98661.1 AJC98662.1 AJC98663.1 AJC98669.1 AJC98673.1 AJC98675.1 KP274514 KP274520 KP274522 KP274525 AJC98660.1 AJC98666.1 AJC98668.1 AJC98671.1 KP274518 AJC98664.1 KP274528 AJC98674.1 KP274526 AJC98672.1 AAAB01008839 EAA05929.4 APCN01004558 JXUM01025687 JXUM01025688 JXUM01025689 JXUM01025690 KQ560730 KXJ81109.1 KPJ11221.1 DS231844 EDS35755.1 GG666563 EEN54998.1 KPI92735.1 CH933807 EDW12821.1 KQ971338 EFA02221.1 JXUM01026933 KQ560772 KXJ80946.1 OUUW01000004 SPP80050.1 CH902624 EDV33327.1 UFQT01000108 SSX20101.1 CH480820 EDW54267.1 CM000361 CM002910 EDX03740.1 KMY88114.1 CM000157 EDW87670.1 KMY88115.1 AE014134 BT031346 AAF50983.2 ABY21759.1 CH916372 EDV99237.1 CH954177 EDV57765.1 KQS70110.1 CH379060 EAL34139.2 CP012523 ALC38127.1 CH479187 EDW39800.1 GDHC01004876 JAQ13753.1 ADMH02001023 ETN64382.1 GGFL01000157 MBW64335.1 OWR50506.1 GEZM01013147 GEZM01013146 JAV92718.1 JRES01000186 KNC33506.1 CCAG010016253 GDHC01008368 JAQ10261.1 JRES01000827 KNC28021.1 CH940649 EDW64279.1 ALC38009.1 JXJN01006233 JXJN01006234 CH963852 EDW75550.2 GAMC01018837 JAB87718.1 GAMC01018836 JAB87719.1 EDW87313.2 SPP79320.1 SPP79322.1 SPP79321.1 ODYU01001738 SOQ38347.1 KRG03784.1 EDW13496.1 GBXI01005367 JAD08925.1 PCG71027.1 CH480829 EDW45573.1 CH963857 EDW75702.2 EDV57401.2 EDW64373.2 GDHF01004480 JAI47834.1 EDX03392.1 KMY87561.1 AAX53591.3 BT100297 ACZ06883.1 CH916379 CH916368 EDV94210.1 EDW02760.1 EAL34013.2 GAMC01016896 GAMC01016895 JAB89660.1 CH902620 EDV31349.1 NEVH01006721 PNF37194.1
KPI92734.1 AGBW02009558 OWR50507.1 KQ460883 KPJ11222.1 RSAL01000299 RVE42787.1 ATLV01015001 KE524999 KFB39841.1 KP274524 AJC98670.1 AXCN02000859 AXCM01000195 KP274530 AJC98676.1 KP274521 AJC98667.1 KP274519 AJC98665.1 KP274531 AJC98677.1 KP274532 AJC98678.1 KP274513 KP274515 KP274516 KP274517 KP274523 KP274527 KP274529 AJC98659.1 AJC98661.1 AJC98662.1 AJC98663.1 AJC98669.1 AJC98673.1 AJC98675.1 KP274514 KP274520 KP274522 KP274525 AJC98660.1 AJC98666.1 AJC98668.1 AJC98671.1 KP274518 AJC98664.1 KP274528 AJC98674.1 KP274526 AJC98672.1 AAAB01008839 EAA05929.4 APCN01004558 JXUM01025687 JXUM01025688 JXUM01025689 JXUM01025690 KQ560730 KXJ81109.1 KPJ11221.1 DS231844 EDS35755.1 GG666563 EEN54998.1 KPI92735.1 CH933807 EDW12821.1 KQ971338 EFA02221.1 JXUM01026933 KQ560772 KXJ80946.1 OUUW01000004 SPP80050.1 CH902624 EDV33327.1 UFQT01000108 SSX20101.1 CH480820 EDW54267.1 CM000361 CM002910 EDX03740.1 KMY88114.1 CM000157 EDW87670.1 KMY88115.1 AE014134 BT031346 AAF50983.2 ABY21759.1 CH916372 EDV99237.1 CH954177 EDV57765.1 KQS70110.1 CH379060 EAL34139.2 CP012523 ALC38127.1 CH479187 EDW39800.1 GDHC01004876 JAQ13753.1 ADMH02001023 ETN64382.1 GGFL01000157 MBW64335.1 OWR50506.1 GEZM01013147 GEZM01013146 JAV92718.1 JRES01000186 KNC33506.1 CCAG010016253 GDHC01008368 JAQ10261.1 JRES01000827 KNC28021.1 CH940649 EDW64279.1 ALC38009.1 JXJN01006233 JXJN01006234 CH963852 EDW75550.2 GAMC01018837 JAB87718.1 GAMC01018836 JAB87719.1 EDW87313.2 SPP79320.1 SPP79322.1 SPP79321.1 ODYU01001738 SOQ38347.1 KRG03784.1 EDW13496.1 GBXI01005367 JAD08925.1 PCG71027.1 CH480829 EDW45573.1 CH963857 EDW75702.2 EDV57401.2 EDW64373.2 GDHF01004480 JAI47834.1 EDX03392.1 KMY87561.1 AAX53591.3 BT100297 ACZ06883.1 CH916379 CH916368 EDV94210.1 EDW02760.1 EAL34013.2 GAMC01016896 GAMC01016895 JAB89660.1 CH902620 EDV31349.1 NEVH01006721 PNF37194.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000283053
+ More
UP000075884 UP000075880 UP000030765 UP000075900 UP000075886 UP000075883 UP000007062 UP000075903 UP000069272 UP000076407 UP000075882 UP000075920 UP000075840 UP000069940 UP000249989 UP000075902 UP000075881 UP000075885 UP000002320 UP000001554 UP000192221 UP000009192 UP000007266 UP000268350 UP000007801 UP000001292 UP000000304 UP000002282 UP000000803 UP000001070 UP000008711 UP000001819 UP000092553 UP000008744 UP000000673 UP000037069 UP000092444 UP000008792 UP000092443 UP000092460 UP000007798 UP000095301 UP000235965
UP000075884 UP000075880 UP000030765 UP000075900 UP000075886 UP000075883 UP000007062 UP000075903 UP000069272 UP000076407 UP000075882 UP000075920 UP000075840 UP000069940 UP000249989 UP000075902 UP000075881 UP000075885 UP000002320 UP000001554 UP000192221 UP000009192 UP000007266 UP000268350 UP000007801 UP000001292 UP000000304 UP000002282 UP000000803 UP000001070 UP000008711 UP000001819 UP000092553 UP000008744 UP000000673 UP000037069 UP000092444 UP000008792 UP000092443 UP000092460 UP000007798 UP000095301 UP000235965
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JM02
A0A2A4JGX1
A0A2H1V173
A0A194PJ37
A0A212F9U5
A0A194R0J4
+ More
A0A3S2N6K1 A0A182NEE1 A0A182J736 A0A084VPE7 A0A0D3QDB5 A0A182RGG1 A0A182QG23 A0A182MRE5 A0A0D3QCN6 A0A0D3QCX8 A0A0D3QDB1 A0A0D3QCY8 A0A0D3QCP3 A0A0D3QCM8 A0A0D3QDA7 A0A0D3QDI4 A0A0D3QDJ3 A0A0D3QCY3 Q7QG58 A0A182VGZ9 A0A182F5W4 A0A182X7N4 A0A182L5N5 A0A182VUZ9 A0A182IE33 A0A182H555 A0A194R5S7 A0A182UAU5 A0A182K6S0 A0A182P383 B0W5V2 A0A1S4J637 C3YY39 A0A194PNN6 A0A1W4UIW6 B4KLE0 D2A2M5 A0A182H630 A0A3B0JX29 B3MTZ8 A0A336LT49 B4I3C2 B4Q324 B4NZ06 A0A0J9TFK6 Q9VR25 B4JQQ7 B3N495 Q29LA6 A0A0M4E6D9 B4GQC8 A0A146M1R4 W5JJ33 A0A087Z3M8 A0A212F9T9 A0A1Y1N5J0 A0A0L0CMK1 A0A1B0F9W5 A0A146LR12 A0A0L0C706 B4LUV2 A0A0M3QSZ8 A0A1A9YQX8 A0A1B0AZB3 B4MTW1 W8AST7 W8BD55 B4NW33 A0A3B0JV17 A0A3B0JGE0 A0A3B0K2T3 A0A2H1VBZ7 A0A0Q9XIZ7 B4KIY3 A0A1I8MMA3 A0A0A1XD89 A0A2A4JG26 B4IDE7 B4MVI2 B3N901 B4LVH9 A0A0K8W9X7 B4Q7L2 A0A0J9QU46 Q59E14 D0Z750 B4JAH4 A0A1I8M2C2 Q29LN2 W8ALP2 B3MJD9 A0A2J7R8N1
A0A3S2N6K1 A0A182NEE1 A0A182J736 A0A084VPE7 A0A0D3QDB5 A0A182RGG1 A0A182QG23 A0A182MRE5 A0A0D3QCN6 A0A0D3QCX8 A0A0D3QDB1 A0A0D3QCY8 A0A0D3QCP3 A0A0D3QCM8 A0A0D3QDA7 A0A0D3QDI4 A0A0D3QDJ3 A0A0D3QCY3 Q7QG58 A0A182VGZ9 A0A182F5W4 A0A182X7N4 A0A182L5N5 A0A182VUZ9 A0A182IE33 A0A182H555 A0A194R5S7 A0A182UAU5 A0A182K6S0 A0A182P383 B0W5V2 A0A1S4J637 C3YY39 A0A194PNN6 A0A1W4UIW6 B4KLE0 D2A2M5 A0A182H630 A0A3B0JX29 B3MTZ8 A0A336LT49 B4I3C2 B4Q324 B4NZ06 A0A0J9TFK6 Q9VR25 B4JQQ7 B3N495 Q29LA6 A0A0M4E6D9 B4GQC8 A0A146M1R4 W5JJ33 A0A087Z3M8 A0A212F9T9 A0A1Y1N5J0 A0A0L0CMK1 A0A1B0F9W5 A0A146LR12 A0A0L0C706 B4LUV2 A0A0M3QSZ8 A0A1A9YQX8 A0A1B0AZB3 B4MTW1 W8AST7 W8BD55 B4NW33 A0A3B0JV17 A0A3B0JGE0 A0A3B0K2T3 A0A2H1VBZ7 A0A0Q9XIZ7 B4KIY3 A0A1I8MMA3 A0A0A1XD89 A0A2A4JG26 B4IDE7 B4MVI2 B3N901 B4LVH9 A0A0K8W9X7 B4Q7L2 A0A0J9QU46 Q59E14 D0Z750 B4JAH4 A0A1I8M2C2 Q29LN2 W8ALP2 B3MJD9 A0A2J7R8N1
PDB
1QZ1
E-value=1.65834e-11,
Score=168
Ontologies
GO
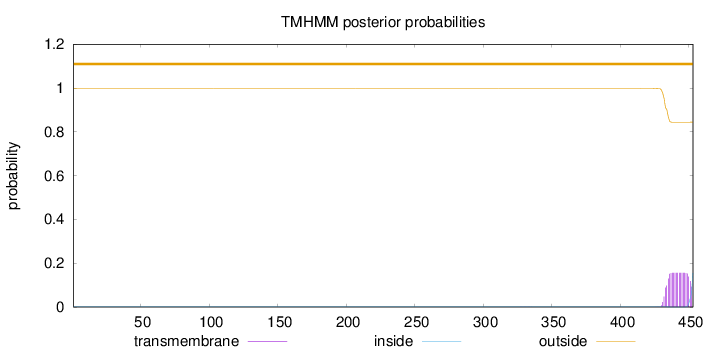
Topology
Length:
453
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.92138
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00110
outside
1 - 453
Population Genetic Test Statistics
Pi
196.171406
Theta
167.527107
Tajima's D
0.224984
CLR
0.426404
CSRT
0.429978501074946
Interpretation
Possibly Positive selection
