Gene
KWMTBOMO07158 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010400
Annotation
triacylglycerol_lipase_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.582
Sequence
CDS
ATGTTGAAACTGATTACTTTGTTGGCCATCGCATTTGCCGCTTGCAACGGAGATGCCATTCAAAACTTGGAGCGATTTGGAAGTTTAAATAAATATTATTACTATAGCAATGCACAGAGAAATTCGATTACACTCACCGAAGACCACTTTCCCACTGGAAACGATACTGCTGCGCCCTTTAACAATAATTCGGATATTGTCGTAATAATACACGGTCACAGTGGCACTGCTACGACGACGATCAACCCTATTGTTAAAGACGCATTTTTAACAAGTGGCGATTATAACGTTATCGTTGTGGATTGGTCATCATTCAGTTTATCAACATATTCAACGGCAGTGATGGCTGTGACAGGTGTGGGATCAAGTATCGCTACTTTTTTGAAAAATCTCAAATTACCACTTAATAAGGTGCACATTGTTGGTTTCAATCTTGGAGCTCATGTTGCTGGAGTGACTGGAAGAAATTTGGAAGGCAAAGTAGCAAGAATTACGGGTCTAGACCCTTCGGCTCGTGATTGGGAAAACAACGTACTACGACTGGGAACAAACGATGCTCAATACGTTGAAGTTATTCACACCGACGGATCTGGTGTCAACAAAAACGGCTTGGGCGTTGCTATTGGACACATTGACTTCTTTGTCAACGGCCGTCTTGTCCAACCGGGCTGCACGAATAACTTATGTAGCCACAACCGCGCTTATGAAGTTTTTGCTGCAACAATTACGCATGGCAAACATTACGGAAATCAGTGCAGCACAGAAGCGGAGATAACCGCAAATAATTGCCGAGGATTTATAGTTGAAATGGGTACTGCCAAACTTGTAAAGCATGGGTATGTTTAA
Protein
MLKLITLLAIAFAACNGDAIQNLERFGSLNKYYYYSNAQRNSITLTEDHFPTGNDTAAPFNNNSDIVVIIHGHSGTATTTINPIVKDAFLTSGDYNVIVVDWSSFSLSTYSTAVMAVTGVGSSIATFLKNLKLPLNKVHIVGFNLGAHVAGVTGRNLEGKVARITGLDPSARDWENNVLRLGTNDAQYVEVIHTDGSGVNKNGLGVAIGHIDFFVNGRLVQPGCTNNLCSHNRAYEVFAATITHGKHYGNQCSTEAEITANNCRGFIVEMGTAKLVKHGYV
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
Q2F644
A0A212EPM5
A0A194PI56
A0A194R0A5
A0A1E1W1Y4
I3QC11
+ More
A9X7K9 B2ZGH8 A0A2A4JCM1 A0A2A4K529 A0A3S2LXV4 A0A194Q4L0 A0A1E1WA09 A0A212FJU4 A0A2A4K3P3 I3QC07 A0A0K8SZM3 A0A0L7KN26 A0A023PMB2 A0A2H1WKE5 A0A2H1V536 A0A0L7LP14 I3QC05 A0A1E1W0R7 B6CMF7 A0A1E1W799 A0A2A4K4B9 A0A2A4K3N9 A0A1E1W907 A0A2H1VFB2 A0A2H1V550 H9IW77 A0A194PYJ4 A0A0N1I7F3 A0A0K8SZQ3 H9IW76 A0A1E1WJS0 A0A2H1V6E0 A0A1E1W9H1 A0A2A4K5B8 A0A194PYJ7 A0A0N1PGL4 A0A1E1WEQ2 A0A2A4K4N6 F5GTG3 A0A0N1PHW4 A0A0N1IP50 A0A1E1VXV5 A0A1E1W2D3 A0A1E1WI38 F6K6Y8 A0A2A4IX28 A0A068B7S7 F5GTF8 F6K6Y9 A0A1E1WKZ4 A0A194PJT7 A0A2H1VUD4 A0A194R1N2 A0A194PW49 A0A194RP46 A0A194PZD7 A0A212F9I1 A0A194R1Y7 A0A068B851 H9IW75 Q6L5Z4 A0A194PVZ0 A0A194PKQ5 A0A2A4JGL3 B6CMF3 Q45EY1 A0A3Q9EEK2 Q8MY66 H9IZI4 A0A212FLK4 A0A2H1VAX5 A0A212FLK2 I3QC12 A0A212FLL3 I3QC04 A0A2A4JGN5 H9IZI5 Q58I81 I1SR90 A0A3S2M9A2 A0A2H1V6Z6 A0A2A4JFT0 Q58I78 A0A2H1WWV2 F5GTG0 A0A194RKD5 A0A068B5W9 A0A2A4JEP6 A0A2A4JFG9 A0A2A4JEN4 A0A2A4JES0
A9X7K9 B2ZGH8 A0A2A4JCM1 A0A2A4K529 A0A3S2LXV4 A0A194Q4L0 A0A1E1WA09 A0A212FJU4 A0A2A4K3P3 I3QC07 A0A0K8SZM3 A0A0L7KN26 A0A023PMB2 A0A2H1WKE5 A0A2H1V536 A0A0L7LP14 I3QC05 A0A1E1W0R7 B6CMF7 A0A1E1W799 A0A2A4K4B9 A0A2A4K3N9 A0A1E1W907 A0A2H1VFB2 A0A2H1V550 H9IW77 A0A194PYJ4 A0A0N1I7F3 A0A0K8SZQ3 H9IW76 A0A1E1WJS0 A0A2H1V6E0 A0A1E1W9H1 A0A2A4K5B8 A0A194PYJ7 A0A0N1PGL4 A0A1E1WEQ2 A0A2A4K4N6 F5GTG3 A0A0N1PHW4 A0A0N1IP50 A0A1E1VXV5 A0A1E1W2D3 A0A1E1WI38 F6K6Y8 A0A2A4IX28 A0A068B7S7 F5GTF8 F6K6Y9 A0A1E1WKZ4 A0A194PJT7 A0A2H1VUD4 A0A194R1N2 A0A194PW49 A0A194RP46 A0A194PZD7 A0A212F9I1 A0A194R1Y7 A0A068B851 H9IW75 Q6L5Z4 A0A194PVZ0 A0A194PKQ5 A0A2A4JGL3 B6CMF3 Q45EY1 A0A3Q9EEK2 Q8MY66 H9IZI4 A0A212FLK4 A0A2H1VAX5 A0A212FLK2 I3QC12 A0A212FLL3 I3QC04 A0A2A4JGN5 H9IZI5 Q58I81 I1SR90 A0A3S2M9A2 A0A2H1V6Z6 A0A2A4JFT0 Q58I78 A0A2H1WWV2 F5GTG0 A0A194RKD5 A0A068B5W9 A0A2A4JEP6 A0A2A4JFG9 A0A2A4JEN4 A0A2A4JES0
EMBL
BABH01005609
DQ311228
ABD36173.1
AGBW02013442
OWR43426.1
KQ459603
+ More
KPI92733.1 KQ460883 KPJ11223.1 GDQN01010826 GDQN01010047 JAT80228.1 JAT81007.1 JF999960 AFI64314.1 DQ875226 ABK29468.1 EU660855 ACD37365.1 NWSH01002009 PCG69448.1 NWSH01000166 PCG78873.1 RSAL01000128 RVE46507.1 KQ459585 KPI98345.1 GDQN01007206 JAT83848.1 AGBW02008213 OWR54002.1 PCG78877.1 JF999956 AFI64310.1 GBRD01007078 JAG58743.1 JTDY01008208 KOB64678.1 KJ439229 AHX25880.1 ODYU01009261 SOQ53555.1 ODYU01000727 SOQ35960.1 JTDY01000484 KOB76966.1 JF999954 AFI64308.1 GDQN01010488 JAT80566.1 EU325557 ACB54947.1 GDQN01008225 JAT82829.1 PCG78876.1 PCG78875.1 GDQN01007598 JAT83456.1 ODYU01002260 SOQ39515.1 SOQ35961.1 BABH01005987 KPI98407.1 KQ460622 KPJ13523.1 GBRD01007079 JAG58742.1 GDQN01003801 JAT87253.1 SOQ35962.1 GDQN01007430 JAT83624.1 PCG78872.1 KPI98406.1 KPJ13527.1 GDQN01005586 JAT85468.1 PCG78874.1 JI484236 AEB26287.1 KPJ13522.1 KPJ13524.1 GDQN01011535 JAT79519.1 GDQN01009916 JAT81138.1 GDQN01004543 JAT86511.1 HM357825 AEA76291.1 NWSH01005705 PCG64026.1 KJ638235 AIC80932.1 JI484231 AEB26282.1 HM357826 AEA76292.1 GDQN01003467 JAT87587.1 KQ459602 KPI93582.1 ODYU01004467 SOQ44366.1 KQ460870 KPJ11708.1 KQ459589 KPI97552.1 KQ460106 KPJ17796.1 KPI98408.1 AGBW02009585 OWR50388.1 KPJ11707.1 KJ638234 AIC80931.1 BABH01005986 AB180932 BAD22559.1 KPI97551.1 KPI93583.1 NWSH01001649 PCG70553.1 EU325553 ACB54943.1 DQ149986 AAZ66799.1 MH246982 AZQ19986.1 DQ286554 AB076385 ABB90967.1 BAC00960.1 BABH01014242 BABH01014243 BABH01014244 BABH01014245 BABH01014246 AGBW02007744 OWR54624.1 ODYU01001581 SOQ38003.1 OWR54623.1 JF999961 AFI64315.1 OWR54622.1 JF999953 AFI64307.1 PCG70552.1 BABH01014247 AY945209 AAX39407.1 JF303743 ADZ54059.1 RSAL01000005 RVE54443.1 ODYU01001007 SOQ36620.1 PCG70558.1 AY945212 EU700252 AAX39410.1 ACD80298.1 ODYU01011603 SOQ57446.1 JI484233 AEB26284.1 KPJ17795.1 KJ638233 AIC80930.1 PCG70555.1 PCG70559.1 PCG70545.1 PCG70557.1
KPI92733.1 KQ460883 KPJ11223.1 GDQN01010826 GDQN01010047 JAT80228.1 JAT81007.1 JF999960 AFI64314.1 DQ875226 ABK29468.1 EU660855 ACD37365.1 NWSH01002009 PCG69448.1 NWSH01000166 PCG78873.1 RSAL01000128 RVE46507.1 KQ459585 KPI98345.1 GDQN01007206 JAT83848.1 AGBW02008213 OWR54002.1 PCG78877.1 JF999956 AFI64310.1 GBRD01007078 JAG58743.1 JTDY01008208 KOB64678.1 KJ439229 AHX25880.1 ODYU01009261 SOQ53555.1 ODYU01000727 SOQ35960.1 JTDY01000484 KOB76966.1 JF999954 AFI64308.1 GDQN01010488 JAT80566.1 EU325557 ACB54947.1 GDQN01008225 JAT82829.1 PCG78876.1 PCG78875.1 GDQN01007598 JAT83456.1 ODYU01002260 SOQ39515.1 SOQ35961.1 BABH01005987 KPI98407.1 KQ460622 KPJ13523.1 GBRD01007079 JAG58742.1 GDQN01003801 JAT87253.1 SOQ35962.1 GDQN01007430 JAT83624.1 PCG78872.1 KPI98406.1 KPJ13527.1 GDQN01005586 JAT85468.1 PCG78874.1 JI484236 AEB26287.1 KPJ13522.1 KPJ13524.1 GDQN01011535 JAT79519.1 GDQN01009916 JAT81138.1 GDQN01004543 JAT86511.1 HM357825 AEA76291.1 NWSH01005705 PCG64026.1 KJ638235 AIC80932.1 JI484231 AEB26282.1 HM357826 AEA76292.1 GDQN01003467 JAT87587.1 KQ459602 KPI93582.1 ODYU01004467 SOQ44366.1 KQ460870 KPJ11708.1 KQ459589 KPI97552.1 KQ460106 KPJ17796.1 KPI98408.1 AGBW02009585 OWR50388.1 KPJ11707.1 KJ638234 AIC80931.1 BABH01005986 AB180932 BAD22559.1 KPI97551.1 KPI93583.1 NWSH01001649 PCG70553.1 EU325553 ACB54943.1 DQ149986 AAZ66799.1 MH246982 AZQ19986.1 DQ286554 AB076385 ABB90967.1 BAC00960.1 BABH01014242 BABH01014243 BABH01014244 BABH01014245 BABH01014246 AGBW02007744 OWR54624.1 ODYU01001581 SOQ38003.1 OWR54623.1 JF999961 AFI64315.1 OWR54622.1 JF999953 AFI64307.1 PCG70552.1 BABH01014247 AY945209 AAX39407.1 JF303743 ADZ54059.1 RSAL01000005 RVE54443.1 ODYU01001007 SOQ36620.1 PCG70558.1 AY945212 EU700252 AAX39410.1 ACD80298.1 ODYU01011603 SOQ57446.1 JI484233 AEB26284.1 KPJ17795.1 KJ638233 AIC80930.1 PCG70555.1 PCG70559.1 PCG70545.1 PCG70557.1
Proteomes
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
Q2F644
A0A212EPM5
A0A194PI56
A0A194R0A5
A0A1E1W1Y4
I3QC11
+ More
A9X7K9 B2ZGH8 A0A2A4JCM1 A0A2A4K529 A0A3S2LXV4 A0A194Q4L0 A0A1E1WA09 A0A212FJU4 A0A2A4K3P3 I3QC07 A0A0K8SZM3 A0A0L7KN26 A0A023PMB2 A0A2H1WKE5 A0A2H1V536 A0A0L7LP14 I3QC05 A0A1E1W0R7 B6CMF7 A0A1E1W799 A0A2A4K4B9 A0A2A4K3N9 A0A1E1W907 A0A2H1VFB2 A0A2H1V550 H9IW77 A0A194PYJ4 A0A0N1I7F3 A0A0K8SZQ3 H9IW76 A0A1E1WJS0 A0A2H1V6E0 A0A1E1W9H1 A0A2A4K5B8 A0A194PYJ7 A0A0N1PGL4 A0A1E1WEQ2 A0A2A4K4N6 F5GTG3 A0A0N1PHW4 A0A0N1IP50 A0A1E1VXV5 A0A1E1W2D3 A0A1E1WI38 F6K6Y8 A0A2A4IX28 A0A068B7S7 F5GTF8 F6K6Y9 A0A1E1WKZ4 A0A194PJT7 A0A2H1VUD4 A0A194R1N2 A0A194PW49 A0A194RP46 A0A194PZD7 A0A212F9I1 A0A194R1Y7 A0A068B851 H9IW75 Q6L5Z4 A0A194PVZ0 A0A194PKQ5 A0A2A4JGL3 B6CMF3 Q45EY1 A0A3Q9EEK2 Q8MY66 H9IZI4 A0A212FLK4 A0A2H1VAX5 A0A212FLK2 I3QC12 A0A212FLL3 I3QC04 A0A2A4JGN5 H9IZI5 Q58I81 I1SR90 A0A3S2M9A2 A0A2H1V6Z6 A0A2A4JFT0 Q58I78 A0A2H1WWV2 F5GTG0 A0A194RKD5 A0A068B5W9 A0A2A4JEP6 A0A2A4JFG9 A0A2A4JEN4 A0A2A4JES0
A9X7K9 B2ZGH8 A0A2A4JCM1 A0A2A4K529 A0A3S2LXV4 A0A194Q4L0 A0A1E1WA09 A0A212FJU4 A0A2A4K3P3 I3QC07 A0A0K8SZM3 A0A0L7KN26 A0A023PMB2 A0A2H1WKE5 A0A2H1V536 A0A0L7LP14 I3QC05 A0A1E1W0R7 B6CMF7 A0A1E1W799 A0A2A4K4B9 A0A2A4K3N9 A0A1E1W907 A0A2H1VFB2 A0A2H1V550 H9IW77 A0A194PYJ4 A0A0N1I7F3 A0A0K8SZQ3 H9IW76 A0A1E1WJS0 A0A2H1V6E0 A0A1E1W9H1 A0A2A4K5B8 A0A194PYJ7 A0A0N1PGL4 A0A1E1WEQ2 A0A2A4K4N6 F5GTG3 A0A0N1PHW4 A0A0N1IP50 A0A1E1VXV5 A0A1E1W2D3 A0A1E1WI38 F6K6Y8 A0A2A4IX28 A0A068B7S7 F5GTF8 F6K6Y9 A0A1E1WKZ4 A0A194PJT7 A0A2H1VUD4 A0A194R1N2 A0A194PW49 A0A194RP46 A0A194PZD7 A0A212F9I1 A0A194R1Y7 A0A068B851 H9IW75 Q6L5Z4 A0A194PVZ0 A0A194PKQ5 A0A2A4JGL3 B6CMF3 Q45EY1 A0A3Q9EEK2 Q8MY66 H9IZI4 A0A212FLK4 A0A2H1VAX5 A0A212FLK2 I3QC12 A0A212FLL3 I3QC04 A0A2A4JGN5 H9IZI5 Q58I81 I1SR90 A0A3S2M9A2 A0A2H1V6Z6 A0A2A4JFT0 Q58I78 A0A2H1WWV2 F5GTG0 A0A194RKD5 A0A068B5W9 A0A2A4JEP6 A0A2A4JFG9 A0A2A4JEN4 A0A2A4JES0
PDB
1GPL
E-value=2.64605e-28,
Score=310
Ontologies
PANTHER
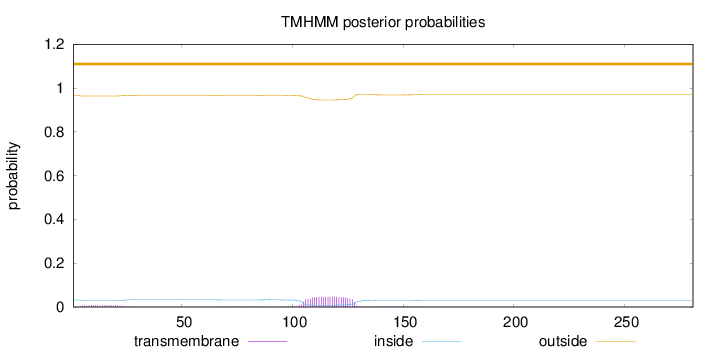
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 17,
Likelihood: 0.996209
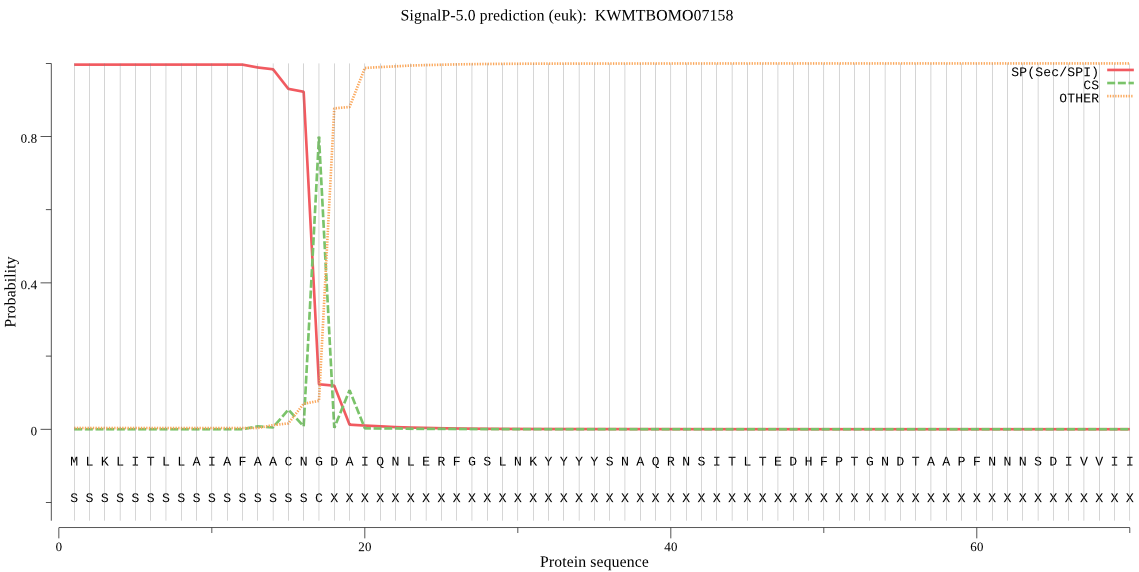
Length:
281
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.28566
Exp number, first 60 AAs:
0.15199
Total prob of N-in:
0.03285
outside
1 - 281
Population Genetic Test Statistics
Pi
198.820219
Theta
115.390298
Tajima's D
1.474674
CLR
19.460605
CSRT
0.785210739463027
Interpretation
Uncertain
