Gene
KWMTBOMO07148
Annotation
PREDICTED:_phospholipase_A1_1_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.149
Sequence
CDS
ATGTATATAGGTTTAGATCCAGCCCGTCCATTGGTGTCTGCGTATGGCTCCCGGCAGTTCCGTCTTAGCAGAGATGACGCTTATGTCGTTCAAGTCTTGCATACCAACGCTGGTTTCCTAGGAGAATCCGGCTTGGTGGGGCACGTGGACTTCTGCATCAACGGTGGACGACTACAGCCGGGCTGCAAAGGACATTTCATGCGTATCGCTCGATGTAGTCATTTCAAGAGCTCGTGTTACTTCGCCGCGAGTGTTCGCAAAAGTCTACGCCTTGTAGGCGTTCCTTGCGACACCTCGTGTCCTAAAGAGCGAGGTCCTTGGGGTATACGGTACGACGGGAAGGCTATCCGGCTTGGGGATGATGTTCCTGATTCGTCTAGAGGAATGTATTGTGTCAAAATAGATCACGAAGAAGATTGTCCTTTCGATTAA
Protein
MYIGLDPARPLVSAYGSRQFRLSRDDAYVVQVLHTNAGFLGESGLVGHVDFCINGGRLQPGCKGHFMRIARCSHFKSSCYFAASVRKSLRLVGVPCDTSCPKERGPWGIRYDGKAIRLGDDVPDSSRGMYCVKIDHEEDCPFD
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A2H1VVD4
A0A3S2LLA2
A0A212FGB1
A0A194PI51
A0A2A4IUC5
A0A0T6BFB5
+ More
U4UHQ7 A0A194R0B0 A0A1W4WZZ2 E0VRR9 A0A067RD55 A0A1B6JLJ0 A0A1B6D9T4 A0A1B6IRW9 A0A2R7WM19 J9K7I3 A0A2H8TMC7 T1H8M0 A0A2J7R8F3 A0A2S2P627 A0A0Q9XFI2 Q173Z3 A0A0L7KLR7 B4MWN9 A0A1A9WZW6 B4KKP2 A0A1S4FFC6 A0A1A9UMK0 B4LUI3 A0A0K8V585 A0A3B0K1L3 A0A0L0BZD9 B3MJU6 A0A1A9XZJ2 A0A0R3NU97 Q29KQ9 A0A0Q5WAV3 A0A0R1DJN8 A0A1W4W402 A0A1I8MSU9 A0A1J1IXA1 A0A0M4EGE6 A0A1S3DUB1 A0A0J9QZV0 A0A0J9R0F0 Q9VKR3 M9MRT1 B3N546 B4P0T3 A0A182PDD4 W8C942 A0A0A1XM29 W8BRU6 A0A182TB02 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182YFE8 A0A182UUE9 A0A182MMH0 A0A182R9Q0 A0A182TKD0 A0A084VCS6 A0A1S4H064 A0A1I8NM26 A0A182GWJ7 B4GSZ1 B4JE61 B4Q9S5 B4HWU4 B0W8V5 A0A1B0CVI9 A0A182JCK0 T1GFZ4 A0A182F5H1 A0A2H8TJ53 A0A2S2PDY1 A0A182KFF2 A0A1B0FLL2 A0A1A9Z7T9 J9JKF5 A0A1B0B900 A0A182WGZ1 A0A182NEW7 A0A2S2Q5X0 A0A336LN77 A0A336K9X1 A0A182MBT5 A0A182GWJ6 A0A067RF91 A0A2J7R8E4 D6X109 T1GFZ3 Q8ITU8 A0A182YFE9 A0A182QP58 Q173Z4
U4UHQ7 A0A194R0B0 A0A1W4WZZ2 E0VRR9 A0A067RD55 A0A1B6JLJ0 A0A1B6D9T4 A0A1B6IRW9 A0A2R7WM19 J9K7I3 A0A2H8TMC7 T1H8M0 A0A2J7R8F3 A0A2S2P627 A0A0Q9XFI2 Q173Z3 A0A0L7KLR7 B4MWN9 A0A1A9WZW6 B4KKP2 A0A1S4FFC6 A0A1A9UMK0 B4LUI3 A0A0K8V585 A0A3B0K1L3 A0A0L0BZD9 B3MJU6 A0A1A9XZJ2 A0A0R3NU97 Q29KQ9 A0A0Q5WAV3 A0A0R1DJN8 A0A1W4W402 A0A1I8MSU9 A0A1J1IXA1 A0A0M4EGE6 A0A1S3DUB1 A0A0J9QZV0 A0A0J9R0F0 Q9VKR3 M9MRT1 B3N546 B4P0T3 A0A182PDD4 W8C942 A0A0A1XM29 W8BRU6 A0A182TB02 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182YFE8 A0A182UUE9 A0A182MMH0 A0A182R9Q0 A0A182TKD0 A0A084VCS6 A0A1S4H064 A0A1I8NM26 A0A182GWJ7 B4GSZ1 B4JE61 B4Q9S5 B4HWU4 B0W8V5 A0A1B0CVI9 A0A182JCK0 T1GFZ4 A0A182F5H1 A0A2H8TJ53 A0A2S2PDY1 A0A182KFF2 A0A1B0FLL2 A0A1A9Z7T9 J9JKF5 A0A1B0B900 A0A182WGZ1 A0A182NEW7 A0A2S2Q5X0 A0A336LN77 A0A336K9X1 A0A182MBT5 A0A182GWJ6 A0A067RF91 A0A2J7R8E4 D6X109 T1GFZ3 Q8ITU8 A0A182YFE9 A0A182QP58 Q173Z4
Pubmed
EMBL
ODYU01004657
SOQ44773.1
RSAL01000070
RVE49156.1
AGBW02008689
OWR52768.1
+ More
KQ459603 KPI92728.1 NWSH01006218 PCG63587.1 LJIG01000941 KRT85999.1 KB632361 ERL93529.1 KQ460883 KPJ11228.1 DS235494 EEB16075.1 KK852543 KDR21672.1 GECU01007744 JAS99962.1 GEDC01014844 JAS22454.1 GECU01018020 JAS89686.1 KK855069 PTY20664.1 ABLF02030156 GFXV01003509 MBW15314.1 ACPB03007015 NEVH01006721 PNF37114.1 GGMR01012271 MBY24890.1 CH933807 KRG03390.1 CH477415 EAT41377.1 JTDY01008980 KOB64242.1 CH963857 EDW76528.1 EDW12706.2 CH940649 EDW64169.2 GDHF01018225 JAI34089.1 OUUW01000006 SPP81760.1 JRES01001119 KNC25385.1 CH902620 EDV32401.1 CH379061 KRT04753.1 EAL33115.2 CH954177 KQS70593.1 CM000157 KRJ97515.1 CVRI01000063 CRL04728.1 CP012523 ALC39767.1 CM002910 KMY89607.1 KMY89606.1 AE014134 BT072821 AAF52997.3 ACN62424.1 ADV37040.1 EDV58991.1 EDW87978.2 GAMC01002484 JAC04072.1 GBXI01001928 JAD12364.1 GAMC01002485 JAC04071.1 AXCN02000839 APCN01003737 AXCM01005154 ATLV01010847 KE524620 KFB35770.1 AAAB01008984 JXUM01093483 KQ564059 KXJ72857.1 CH479189 EDW25500.1 CH916368 EDW03581.1 CM000361 EDX04592.1 CH480818 EDW52489.1 DS231860 EDS39334.1 AJWK01030623 CAQQ02088126 CAQQ02088127 CAQQ02088128 CAQQ02088129 GFXV01001513 MBW13318.1 GGMR01014986 MBY27605.1 CCAG010020502 ABLF02013799 JXJN01010172 JXJN01010173 JXJN01010174 JXJN01010175 JXJN01010176 GGMS01003952 MBY73155.1 UFQT01000031 SSX18231.1 UFQS01000230 UFQT01000230 SSX01714.1 SSX22094.1 JXUM01093479 JXUM01093480 JXUM01093481 JXUM01093482 KXJ72856.1 KDR21673.1 PNF37113.1 KQ971371 EFA09985.1 CAQQ02088132 AF303984 AAN31169.1 EAT41376.1
KQ459603 KPI92728.1 NWSH01006218 PCG63587.1 LJIG01000941 KRT85999.1 KB632361 ERL93529.1 KQ460883 KPJ11228.1 DS235494 EEB16075.1 KK852543 KDR21672.1 GECU01007744 JAS99962.1 GEDC01014844 JAS22454.1 GECU01018020 JAS89686.1 KK855069 PTY20664.1 ABLF02030156 GFXV01003509 MBW15314.1 ACPB03007015 NEVH01006721 PNF37114.1 GGMR01012271 MBY24890.1 CH933807 KRG03390.1 CH477415 EAT41377.1 JTDY01008980 KOB64242.1 CH963857 EDW76528.1 EDW12706.2 CH940649 EDW64169.2 GDHF01018225 JAI34089.1 OUUW01000006 SPP81760.1 JRES01001119 KNC25385.1 CH902620 EDV32401.1 CH379061 KRT04753.1 EAL33115.2 CH954177 KQS70593.1 CM000157 KRJ97515.1 CVRI01000063 CRL04728.1 CP012523 ALC39767.1 CM002910 KMY89607.1 KMY89606.1 AE014134 BT072821 AAF52997.3 ACN62424.1 ADV37040.1 EDV58991.1 EDW87978.2 GAMC01002484 JAC04072.1 GBXI01001928 JAD12364.1 GAMC01002485 JAC04071.1 AXCN02000839 APCN01003737 AXCM01005154 ATLV01010847 KE524620 KFB35770.1 AAAB01008984 JXUM01093483 KQ564059 KXJ72857.1 CH479189 EDW25500.1 CH916368 EDW03581.1 CM000361 EDX04592.1 CH480818 EDW52489.1 DS231860 EDS39334.1 AJWK01030623 CAQQ02088126 CAQQ02088127 CAQQ02088128 CAQQ02088129 GFXV01001513 MBW13318.1 GGMR01014986 MBY27605.1 CCAG010020502 ABLF02013799 JXJN01010172 JXJN01010173 JXJN01010174 JXJN01010175 JXJN01010176 GGMS01003952 MBY73155.1 UFQT01000031 SSX18231.1 UFQS01000230 UFQT01000230 SSX01714.1 SSX22094.1 JXUM01093479 JXUM01093480 JXUM01093481 JXUM01093482 KXJ72856.1 KDR21673.1 PNF37113.1 KQ971371 EFA09985.1 CAQQ02088132 AF303984 AAN31169.1 EAT41376.1
Proteomes
UP000283053
UP000007151
UP000053268
UP000218220
UP000030742
UP000053240
+ More
UP000192223 UP000009046 UP000027135 UP000007819 UP000015103 UP000235965 UP000009192 UP000008820 UP000037510 UP000007798 UP000091820 UP000078200 UP000008792 UP000268350 UP000037069 UP000007801 UP000092443 UP000001819 UP000008711 UP000002282 UP000192221 UP000095301 UP000183832 UP000092553 UP000079169 UP000000803 UP000075885 UP000075901 UP000075886 UP000076407 UP000075840 UP000076408 UP000075903 UP000075883 UP000075900 UP000075902 UP000030765 UP000095300 UP000069940 UP000249989 UP000008744 UP000001070 UP000000304 UP000001292 UP000002320 UP000092461 UP000075880 UP000015102 UP000069272 UP000075881 UP000092444 UP000092445 UP000092460 UP000075920 UP000075884 UP000007266
UP000192223 UP000009046 UP000027135 UP000007819 UP000015103 UP000235965 UP000009192 UP000008820 UP000037510 UP000007798 UP000091820 UP000078200 UP000008792 UP000268350 UP000037069 UP000007801 UP000092443 UP000001819 UP000008711 UP000002282 UP000192221 UP000095301 UP000183832 UP000092553 UP000079169 UP000000803 UP000075885 UP000075901 UP000075886 UP000076407 UP000075840 UP000076408 UP000075903 UP000075883 UP000075900 UP000075902 UP000030765 UP000095300 UP000069940 UP000249989 UP000008744 UP000001070 UP000000304 UP000001292 UP000002320 UP000092461 UP000075880 UP000015102 UP000069272 UP000075881 UP000092444 UP000092445 UP000092460 UP000075920 UP000075884 UP000007266
PRIDE
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2H1VVD4
A0A3S2LLA2
A0A212FGB1
A0A194PI51
A0A2A4IUC5
A0A0T6BFB5
+ More
U4UHQ7 A0A194R0B0 A0A1W4WZZ2 E0VRR9 A0A067RD55 A0A1B6JLJ0 A0A1B6D9T4 A0A1B6IRW9 A0A2R7WM19 J9K7I3 A0A2H8TMC7 T1H8M0 A0A2J7R8F3 A0A2S2P627 A0A0Q9XFI2 Q173Z3 A0A0L7KLR7 B4MWN9 A0A1A9WZW6 B4KKP2 A0A1S4FFC6 A0A1A9UMK0 B4LUI3 A0A0K8V585 A0A3B0K1L3 A0A0L0BZD9 B3MJU6 A0A1A9XZJ2 A0A0R3NU97 Q29KQ9 A0A0Q5WAV3 A0A0R1DJN8 A0A1W4W402 A0A1I8MSU9 A0A1J1IXA1 A0A0M4EGE6 A0A1S3DUB1 A0A0J9QZV0 A0A0J9R0F0 Q9VKR3 M9MRT1 B3N546 B4P0T3 A0A182PDD4 W8C942 A0A0A1XM29 W8BRU6 A0A182TB02 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182YFE8 A0A182UUE9 A0A182MMH0 A0A182R9Q0 A0A182TKD0 A0A084VCS6 A0A1S4H064 A0A1I8NM26 A0A182GWJ7 B4GSZ1 B4JE61 B4Q9S5 B4HWU4 B0W8V5 A0A1B0CVI9 A0A182JCK0 T1GFZ4 A0A182F5H1 A0A2H8TJ53 A0A2S2PDY1 A0A182KFF2 A0A1B0FLL2 A0A1A9Z7T9 J9JKF5 A0A1B0B900 A0A182WGZ1 A0A182NEW7 A0A2S2Q5X0 A0A336LN77 A0A336K9X1 A0A182MBT5 A0A182GWJ6 A0A067RF91 A0A2J7R8E4 D6X109 T1GFZ3 Q8ITU8 A0A182YFE9 A0A182QP58 Q173Z4
U4UHQ7 A0A194R0B0 A0A1W4WZZ2 E0VRR9 A0A067RD55 A0A1B6JLJ0 A0A1B6D9T4 A0A1B6IRW9 A0A2R7WM19 J9K7I3 A0A2H8TMC7 T1H8M0 A0A2J7R8F3 A0A2S2P627 A0A0Q9XFI2 Q173Z3 A0A0L7KLR7 B4MWN9 A0A1A9WZW6 B4KKP2 A0A1S4FFC6 A0A1A9UMK0 B4LUI3 A0A0K8V585 A0A3B0K1L3 A0A0L0BZD9 B3MJU6 A0A1A9XZJ2 A0A0R3NU97 Q29KQ9 A0A0Q5WAV3 A0A0R1DJN8 A0A1W4W402 A0A1I8MSU9 A0A1J1IXA1 A0A0M4EGE6 A0A1S3DUB1 A0A0J9QZV0 A0A0J9R0F0 Q9VKR3 M9MRT1 B3N546 B4P0T3 A0A182PDD4 W8C942 A0A0A1XM29 W8BRU6 A0A182TB02 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182YFE8 A0A182UUE9 A0A182MMH0 A0A182R9Q0 A0A182TKD0 A0A084VCS6 A0A1S4H064 A0A1I8NM26 A0A182GWJ7 B4GSZ1 B4JE61 B4Q9S5 B4HWU4 B0W8V5 A0A1B0CVI9 A0A182JCK0 T1GFZ4 A0A182F5H1 A0A2H8TJ53 A0A2S2PDY1 A0A182KFF2 A0A1B0FLL2 A0A1A9Z7T9 J9JKF5 A0A1B0B900 A0A182WGZ1 A0A182NEW7 A0A2S2Q5X0 A0A336LN77 A0A336K9X1 A0A182MBT5 A0A182GWJ6 A0A067RF91 A0A2J7R8E4 D6X109 T1GFZ3 Q8ITU8 A0A182YFE9 A0A182QP58 Q173Z4
PDB
1ETH
E-value=3.04413e-10,
Score=150
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
Length:
143
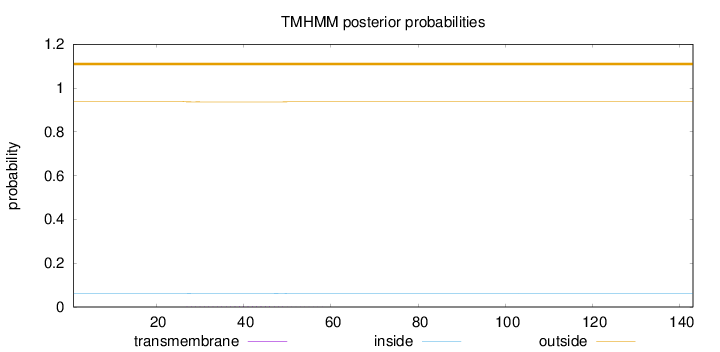
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0545100000000001
Exp number, first 60 AAs:
0.05431
Total prob of N-in:
0.06199
outside
1 - 143
Population Genetic Test Statistics
Pi
304.030322
Theta
188.503803
Tajima's D
1.662368
CLR
0.3068
CSRT
0.823108844557772
Interpretation
Uncertain
