Gene
KWMTBOMO07146 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010403
Annotation
mitochondrial_aldehyde_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.376
Sequence
CDS
ATGGTGAAAGTAGACGTCAAGTACACTAAGCTATTTATCAACAATGAATGGGTAGATGCTGTCAGCAAAAAAACCTTCCCTACCATCAACCCACAAGATGAAACAGTCATAACACAGGTCGCTGAAGGAGATAAGGCGGATATTGATTTGGCCGTCGCCGCAGCAAAGAAAGCATTCCACCGTTATTCCCCATGGCGTACCATGGACGCTTCTCAACGCGGCCTCCTATTGCTTAAGCTAGCCGAACTTCTAGAATCCCAATCTAGATACCTAGCTGAGCTAGAGACTTTAGACTGTGGAAAACCGGTAAAACAATCCGAAGAAGAAGTATACTTTTCCGCAAGCGTGCTCAGATATTACGCTGGAAAAGCGGACAAGATATTAGGAAATACCATTCCTTCAGACGGAGAAGTCCTTACGTTCACCATGAAGGAGCCTGTCGGAGTGTGCGGTCAGATAATACCGTGGAATTACCCCATACCAATGATGTCTTGGAAAATAGCACCAGCCTTAGCTGCAGGATGTACCGTTGTTCTGAAGCCAGCTGAACAAACACCGCTGACTGCGCTCGCTGTTGCCGCCCTGGTCAAAGAAGCGGGCTTTCCACCGGGGGTTGTCAATGTCGTTCCCGGATACGGTGCGACAGCGGGCGGTGCTCTTACTCACCACCCAGATGTTGACAAAATCGCTTTCACTGGCTCCACTGAGGTATACTTTTTTTCTATTGCTTAG
Protein
MVKVDVKYTKLFINNEWVDAVSKKTFPTINPQDETVITQVAEGDKADIDLAVAAAKKAFHRYSPWRTMDASQRGLLLLKLAELLESQSRYLAELETLDCGKPVKQSEEEVYFSASVLRYYAGKADKILGNTIPSDGEVLTFTMKEPVGVCGQIIPWNYPIPMMSWKIAPALAAGCTVVLKPAEQTPLTALAVAALVKEAGFPPGVVNVVPGYGATAGGALTHHPDVDKIAFTGSTEVYFFSIA
Summary
Similarity
Belongs to the aldehyde dehydrogenase family.
Belongs to the protein disulfide isomerase family.
Belongs to the protein disulfide isomerase family.
Uniprot
Q2F5Y9
A0A194PHE0
D2SNR7
A0A194R140
A0A194R1V2
A0A2A4JJH0
+ More
A0A2H1VX41 A0A0L7LPC3 A0A2H1VID4 A0A212EPN8 A0A212EPE8 D2SNQ8 B0X5Q3 A0A2M4AF40 A0A2M4AEX2 A0A1D2N1N3 A0A1S3IR17 A0A1S3HNG6 T1DF46 A0A182QSK4 A0A182GPP9 A0A1J1HJ07 Q16UC5 A0A023EVW3 A0A182F3L2 A0A1S3HRB3 D6X3U7 A0A1Q3FGB5 A0A1S4FNU3 A7XZK3 F6UAT6 W5JT01 A0A1W7R9T2 G3WMK0 A0A182XHT7 A0A182HRD9 A0A369S0Y8 A0A1S4H2G8 Q7Q165 A0A369S1T2 A0A1J1J3B3 B3RSM9 A0A182KYZ4 A0A182VIA5 A0A1B2YLJ7 A0A2U4CG69 A0A384BC88 A1KXI6 F7C0D1 A0A0N8BST4 A0A2S2R0V2 A0A154P517 A0A3G5AQ09 K1QVG5 A0A2F0B3F8 W4XV28 A0A3G5ANS0 K1R266 A0A3S3NWG2 A0A146N6N5 A0A210Q7D8 A0A1S2ZT62 A0A2Y9QA90 A1KXH7 A0A2Y9FAN7 A0A182U026 A0A1W4X864 V4BLL6 L9L452 A0A2R4A6R7 A0A1I8I940 A0A267EC57 A0A1I8JFG3 C3XVV4 A0A2D0PLT6 A0A146XLL6 A0A2K6EL72 E9IRW6 A0A146XJ16 A0A1U8C9A1 G3H7E6 B3RSM8 A0A3B1K1J7 A0A3B1IZU8 G3V7I5 A0A1A6G234 A0A0P5Y166 A0A287CXM9 A0A1S3JX91 A0A087TDX4 E2AFW0 A0A098M1T5 H0Y163 A0A0P4Z8Q6 A0A2S2PZM6 I3N1D1 E2RHQ0 A0A0P4XW28 V5GV38 A0A1S3HPE0
A0A2H1VX41 A0A0L7LPC3 A0A2H1VID4 A0A212EPN8 A0A212EPE8 D2SNQ8 B0X5Q3 A0A2M4AF40 A0A2M4AEX2 A0A1D2N1N3 A0A1S3IR17 A0A1S3HNG6 T1DF46 A0A182QSK4 A0A182GPP9 A0A1J1HJ07 Q16UC5 A0A023EVW3 A0A182F3L2 A0A1S3HRB3 D6X3U7 A0A1Q3FGB5 A0A1S4FNU3 A7XZK3 F6UAT6 W5JT01 A0A1W7R9T2 G3WMK0 A0A182XHT7 A0A182HRD9 A0A369S0Y8 A0A1S4H2G8 Q7Q165 A0A369S1T2 A0A1J1J3B3 B3RSM9 A0A182KYZ4 A0A182VIA5 A0A1B2YLJ7 A0A2U4CG69 A0A384BC88 A1KXI6 F7C0D1 A0A0N8BST4 A0A2S2R0V2 A0A154P517 A0A3G5AQ09 K1QVG5 A0A2F0B3F8 W4XV28 A0A3G5ANS0 K1R266 A0A3S3NWG2 A0A146N6N5 A0A210Q7D8 A0A1S2ZT62 A0A2Y9QA90 A1KXH7 A0A2Y9FAN7 A0A182U026 A0A1W4X864 V4BLL6 L9L452 A0A2R4A6R7 A0A1I8I940 A0A267EC57 A0A1I8JFG3 C3XVV4 A0A2D0PLT6 A0A146XLL6 A0A2K6EL72 E9IRW6 A0A146XJ16 A0A1U8C9A1 G3H7E6 B3RSM8 A0A3B1K1J7 A0A3B1IZU8 G3V7I5 A0A1A6G234 A0A0P5Y166 A0A287CXM9 A0A1S3JX91 A0A087TDX4 E2AFW0 A0A098M1T5 H0Y163 A0A0P4Z8Q6 A0A2S2PZM6 I3N1D1 E2RHQ0 A0A0P4XW28 V5GV38 A0A1S3HPE0
Pubmed
19121390
26354079
20074338
26227816
22118469
27289101
+ More
26383154 26483478 17510324 24945155 18362917 19820115 18464734 20920257 23761445 21709235 30042472 12364791 18719581 20966253 17495919 22992520 28812685 23254933 23385571 29492455 26392545 18563158 21282665 21804562 25329095 15057822 15632090 20798317 16341006
26383154 26483478 17510324 24945155 18362917 19820115 18464734 20920257 23761445 21709235 30042472 12364791 18719581 20966253 17495919 22992520 28812685 23254933 23385571 29492455 26392545 18563158 21282665 21804562 25329095 15057822 15632090 20798317 16341006
EMBL
BABH01005570
BABH01005571
BABH01005572
BABH01005573
DQ311283
ABD36228.1
+ More
KQ459603 KPI92727.1 EZ407197 ACX53758.1 KQ460883 KPJ11229.1 KPJ11230.1 NWSH01001358 PCG71550.1 ODYU01004657 SOQ44774.1 JTDY01000429 KOB77219.1 ODYU01002733 SOQ40599.1 AGBW02013442 OWR43436.1 AGBW02013494 OWR43337.1 EZ407188 ACX53749.1 DS232392 EDS41034.1 GGFK01006088 MBW39409.1 GGFK01006020 MBW39341.1 LJIJ01000295 ODM99179.1 GAMD01003251 JAA98339.1 AXCN02000654 JXUM01078869 KQ563093 KXJ74487.1 CVRI01000006 CRK88023.1 CH477625 EAT38124.1 GAPW01001149 JAC12449.1 KQ971326 EEZ97369.1 GFDL01008464 JAV26581.1 EU106624 ABU97475.1 ADMH02000539 ETN66060.1 GFAH01000487 JAV47902.1 AEFK01074650 APCN01001694 NOWV01000103 RDD40389.1 AAAB01008980 EAA14068.3 RDD40374.1 CVRI01000066 CRL06284.1 DS985243 EDV26543.1 KX060612 AOD75396.1 AY291328 AAQ24547.1 GDIQ01148387 JAL03339.1 GGMS01014351 MBY83554.1 KQ434820 KZC07026.1 MH979760 AYV89215.1 JH817012 EKC37583.1 NTJE010061653 MBV97550.1 AAGJ04009886 AAGJ04009887 MH990439 AYV88986.1 EKC37584.1 NCKU01002086 RWS10468.1 GCES01158808 JAQ27514.1 NEDP02004706 OWF44662.1 AY283296 AAP35081.1 KB202567 ESO89599.1 KB320515 ELW69851.1 KY679131 AVR59242.1 NIVC01002449 PAA57759.1 NIVC01002372 PAA58467.1 NIVC01000222 PAA87659.1 GG666469 EEN67897.1 GCES01044211 JAR42112.1 GL765283 EFZ16677.1 GCES01044212 JAR42111.1 JH000194 EGW07916.1 EDV26544.1 AC133299 CH473962 EDL98822.1 LZPO01107946 OBS59865.1 GDIP01066067 JAM37648.1 AGTP01043698 KK114785 KFM63313.1 GL439158 EFN67709.1 GBSI01000206 JAC96289.1 AAQR03033261 GDIP01220392 JAJ03010.1 GGMS01001617 MBY70820.1 AAEX03007973 GDIP01238190 JAI85211.1 GALX01004368 JAB64098.1
KQ459603 KPI92727.1 EZ407197 ACX53758.1 KQ460883 KPJ11229.1 KPJ11230.1 NWSH01001358 PCG71550.1 ODYU01004657 SOQ44774.1 JTDY01000429 KOB77219.1 ODYU01002733 SOQ40599.1 AGBW02013442 OWR43436.1 AGBW02013494 OWR43337.1 EZ407188 ACX53749.1 DS232392 EDS41034.1 GGFK01006088 MBW39409.1 GGFK01006020 MBW39341.1 LJIJ01000295 ODM99179.1 GAMD01003251 JAA98339.1 AXCN02000654 JXUM01078869 KQ563093 KXJ74487.1 CVRI01000006 CRK88023.1 CH477625 EAT38124.1 GAPW01001149 JAC12449.1 KQ971326 EEZ97369.1 GFDL01008464 JAV26581.1 EU106624 ABU97475.1 ADMH02000539 ETN66060.1 GFAH01000487 JAV47902.1 AEFK01074650 APCN01001694 NOWV01000103 RDD40389.1 AAAB01008980 EAA14068.3 RDD40374.1 CVRI01000066 CRL06284.1 DS985243 EDV26543.1 KX060612 AOD75396.1 AY291328 AAQ24547.1 GDIQ01148387 JAL03339.1 GGMS01014351 MBY83554.1 KQ434820 KZC07026.1 MH979760 AYV89215.1 JH817012 EKC37583.1 NTJE010061653 MBV97550.1 AAGJ04009886 AAGJ04009887 MH990439 AYV88986.1 EKC37584.1 NCKU01002086 RWS10468.1 GCES01158808 JAQ27514.1 NEDP02004706 OWF44662.1 AY283296 AAP35081.1 KB202567 ESO89599.1 KB320515 ELW69851.1 KY679131 AVR59242.1 NIVC01002449 PAA57759.1 NIVC01002372 PAA58467.1 NIVC01000222 PAA87659.1 GG666469 EEN67897.1 GCES01044211 JAR42112.1 GL765283 EFZ16677.1 GCES01044212 JAR42111.1 JH000194 EGW07916.1 EDV26544.1 AC133299 CH473962 EDL98822.1 LZPO01107946 OBS59865.1 GDIP01066067 JAM37648.1 AGTP01043698 KK114785 KFM63313.1 GL439158 EFN67709.1 GBSI01000206 JAC96289.1 AAQR03033261 GDIP01220392 JAJ03010.1 GGMS01001617 MBY70820.1 AAEX03007973 GDIP01238190 JAI85211.1 GALX01004368 JAB64098.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000037510
UP000007151
+ More
UP000002320 UP000094527 UP000085678 UP000075886 UP000069940 UP000249989 UP000183832 UP000008820 UP000069272 UP000007266 UP000002279 UP000000673 UP000007648 UP000076407 UP000075840 UP000253843 UP000007062 UP000009022 UP000075882 UP000075903 UP000245320 UP000261681 UP000002280 UP000076502 UP000005408 UP000007110 UP000285301 UP000242188 UP000079721 UP000248483 UP000248484 UP000075902 UP000192223 UP000030746 UP000011518 UP000095280 UP000215902 UP000001554 UP000221080 UP000233160 UP000189706 UP000001075 UP000018467 UP000002494 UP000092124 UP000005215 UP000054359 UP000000311 UP000005225 UP000002254
UP000002320 UP000094527 UP000085678 UP000075886 UP000069940 UP000249989 UP000183832 UP000008820 UP000069272 UP000007266 UP000002279 UP000000673 UP000007648 UP000076407 UP000075840 UP000253843 UP000007062 UP000009022 UP000075882 UP000075903 UP000245320 UP000261681 UP000002280 UP000076502 UP000005408 UP000007110 UP000285301 UP000242188 UP000079721 UP000248483 UP000248484 UP000075902 UP000192223 UP000030746 UP000011518 UP000095280 UP000215902 UP000001554 UP000221080 UP000233160 UP000189706 UP000001075 UP000018467 UP000002494 UP000092124 UP000005215 UP000054359 UP000000311 UP000005225 UP000002254
Interpro
Gene 3D
ProteinModelPortal
Q2F5Y9
A0A194PHE0
D2SNR7
A0A194R140
A0A194R1V2
A0A2A4JJH0
+ More
A0A2H1VX41 A0A0L7LPC3 A0A2H1VID4 A0A212EPN8 A0A212EPE8 D2SNQ8 B0X5Q3 A0A2M4AF40 A0A2M4AEX2 A0A1D2N1N3 A0A1S3IR17 A0A1S3HNG6 T1DF46 A0A182QSK4 A0A182GPP9 A0A1J1HJ07 Q16UC5 A0A023EVW3 A0A182F3L2 A0A1S3HRB3 D6X3U7 A0A1Q3FGB5 A0A1S4FNU3 A7XZK3 F6UAT6 W5JT01 A0A1W7R9T2 G3WMK0 A0A182XHT7 A0A182HRD9 A0A369S0Y8 A0A1S4H2G8 Q7Q165 A0A369S1T2 A0A1J1J3B3 B3RSM9 A0A182KYZ4 A0A182VIA5 A0A1B2YLJ7 A0A2U4CG69 A0A384BC88 A1KXI6 F7C0D1 A0A0N8BST4 A0A2S2R0V2 A0A154P517 A0A3G5AQ09 K1QVG5 A0A2F0B3F8 W4XV28 A0A3G5ANS0 K1R266 A0A3S3NWG2 A0A146N6N5 A0A210Q7D8 A0A1S2ZT62 A0A2Y9QA90 A1KXH7 A0A2Y9FAN7 A0A182U026 A0A1W4X864 V4BLL6 L9L452 A0A2R4A6R7 A0A1I8I940 A0A267EC57 A0A1I8JFG3 C3XVV4 A0A2D0PLT6 A0A146XLL6 A0A2K6EL72 E9IRW6 A0A146XJ16 A0A1U8C9A1 G3H7E6 B3RSM8 A0A3B1K1J7 A0A3B1IZU8 G3V7I5 A0A1A6G234 A0A0P5Y166 A0A287CXM9 A0A1S3JX91 A0A087TDX4 E2AFW0 A0A098M1T5 H0Y163 A0A0P4Z8Q6 A0A2S2PZM6 I3N1D1 E2RHQ0 A0A0P4XW28 V5GV38 A0A1S3HPE0
A0A2H1VX41 A0A0L7LPC3 A0A2H1VID4 A0A212EPN8 A0A212EPE8 D2SNQ8 B0X5Q3 A0A2M4AF40 A0A2M4AEX2 A0A1D2N1N3 A0A1S3IR17 A0A1S3HNG6 T1DF46 A0A182QSK4 A0A182GPP9 A0A1J1HJ07 Q16UC5 A0A023EVW3 A0A182F3L2 A0A1S3HRB3 D6X3U7 A0A1Q3FGB5 A0A1S4FNU3 A7XZK3 F6UAT6 W5JT01 A0A1W7R9T2 G3WMK0 A0A182XHT7 A0A182HRD9 A0A369S0Y8 A0A1S4H2G8 Q7Q165 A0A369S1T2 A0A1J1J3B3 B3RSM9 A0A182KYZ4 A0A182VIA5 A0A1B2YLJ7 A0A2U4CG69 A0A384BC88 A1KXI6 F7C0D1 A0A0N8BST4 A0A2S2R0V2 A0A154P517 A0A3G5AQ09 K1QVG5 A0A2F0B3F8 W4XV28 A0A3G5ANS0 K1R266 A0A3S3NWG2 A0A146N6N5 A0A210Q7D8 A0A1S2ZT62 A0A2Y9QA90 A1KXH7 A0A2Y9FAN7 A0A182U026 A0A1W4X864 V4BLL6 L9L452 A0A2R4A6R7 A0A1I8I940 A0A267EC57 A0A1I8JFG3 C3XVV4 A0A2D0PLT6 A0A146XLL6 A0A2K6EL72 E9IRW6 A0A146XJ16 A0A1U8C9A1 G3H7E6 B3RSM8 A0A3B1K1J7 A0A3B1IZU8 G3V7I5 A0A1A6G234 A0A0P5Y166 A0A287CXM9 A0A1S3JX91 A0A087TDX4 E2AFW0 A0A098M1T5 H0Y163 A0A0P4Z8Q6 A0A2S2PZM6 I3N1D1 E2RHQ0 A0A0P4XW28 V5GV38 A0A1S3HPE0
PDB
1AG8
E-value=1.50898e-62,
Score=605
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00981 Insect hormone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00340 Histidine metabolism - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00981 Insect hormone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
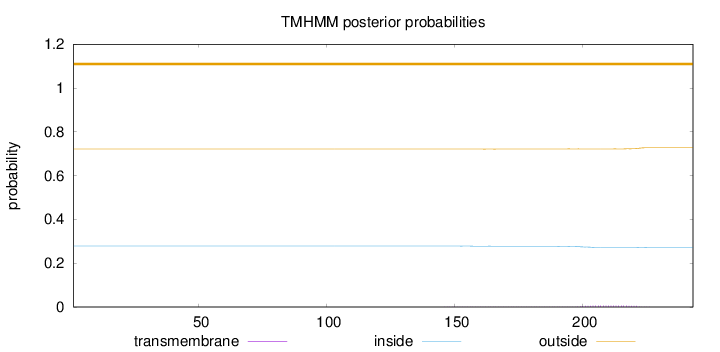
Topology
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18131
Exp number, first 60 AAs:
0
Total prob of N-in:
0.27851
outside
1 - 243
Population Genetic Test Statistics
Pi
215.558087
Theta
165.10144
Tajima's D
0.502949
CLR
0.837497
CSRT
0.512624368781561
Interpretation
Uncertain
