Gene
KWMTBOMO07144
Annotation
PREDICTED:_hepatic_triacylglycerol_lipase-like_[Plutella_xylostella]
Location in the cell
Lysosomal Reliability : 1.051 Mitochondrial Reliability : 1.276
Sequence
CDS
ATGGATTTTCGATATTGGCGATGCGTCATGAAAAGAGAATGGAAATGTCCTGATAGCGAAATTAACTTTTTTTTATATACACCAGAGCATCCATTCAAGCAGTGGGTCGACGTTCGACATGGCGTAGATCACCTGTACGCGTACGGCTGGAAGCCTGACCGAAAAAATGTAATGATCATTCACGGCTTCAACGGAACGTACTCGAAGAGTCCAATGACATTCATACGTGACGCTTACTTGTCTCGCAAGGACTACAACGTGTTCATGGTGGACTGGGCTCCGTTAACGAGGTTTCCGTGTTACCTCTCATCGCTTTCGAACTTGAAATTGGCTGCTCAATGTACTGCTCAGCTTTACTCGTACTTGACGCAAGGGGGCGCTTTGGCAAAGATGATAACTTGCGTGGGGCATTCGTTGGGGGCTCACGTCTGCGGGATGATGTCTAACCACCTAACCGAGAAGCAGTACAAAATTGTAGATTAG
Protein
MDFRYWRCVMKREWKCPDSEINFFLYTPEHPFKQWVDVRHGVDHLYAYGWKPDRKNVMIIHGFNGTYSKSPMTFIRDAYLSRKDYNVFMVDWAPLTRFPCYLSSLSNLKLAAQCTAQLYSYLTQGGALAKMITCVGHSLGAHVCGMMSNHLTEKQYKIVD
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A212FGA9
A0A194R5T7
A0A194PNM6
A0A2H1VVD4
A0A2A4IYV3
A0A2H1VKU5
+ More
X1WKQ9 J9K7I3 A0A2H8TMC7 A0A2S2P627 E0VRR9 A0A1B6IRW9 T1H8M0 A0A2J7R8F3 D7EK20 A0A067RD55 A0A1B6D9T4 N6TUH5 A0A1S3DUB1 J9JKF5 A0A2P8XP77 A0A2H8TJ53 A0A182MMH0 A0A182R9Q0 A0A1S4FFC6 A0A084VCS6 A0A182YFE8 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182JCK0 A0A2R7WM19 B0W8V5 A0A1B6JLJ0 A0A182F5H1 A0A182NEW7 A0A1W4W402 B4HWU4 A0A0J9QZV0 Q9VKR3 A0A1J1IXA1 B3N546 B3MJU6 B4GSZ1 B4P0T3 A0A3B0K1L3 Q29KQ9 A0A0L0BZD9 B4JE61 B4MWN9 A0A2S2PDY1 A0A0M4EGE6 A0A1A9XZJ2 W5JCY0 A0A1A9WZW6 B4KKP2 A0A1I8NM26 A0A1B0B900 A0A1I8MSU9 A0A336LN77 A0A0K8V585 Q7PW37 W8C942 A0A0R3NU97 A0A2S2Q5X0 Q173Z3 W8BRU6 A0A0J9R0F0 A0A182PDD4 M9MRT1 A0A0A1XM29 B4LUI3 A0A182KFF2 B4Q9S5 T1GFZ4 A0A1A9Z7T9 A0A1B0FLL2 A0A1A9UMK0 A0A182WGZ1 A0A1B0CVI9 A0A0N1PHU7 A0A212FF39 A0A2A4IV85 A0A194Q079 A0A3S2M6M2 A0A1J1IWQ7 A0A1B6EGK5 A0A0C9R6H2 K7IRS5 A0A1S3DMV9 A0A232F8T8 A0A195D3J4 A0A158NQR1 A0A195BEA4
X1WKQ9 J9K7I3 A0A2H8TMC7 A0A2S2P627 E0VRR9 A0A1B6IRW9 T1H8M0 A0A2J7R8F3 D7EK20 A0A067RD55 A0A1B6D9T4 N6TUH5 A0A1S3DUB1 J9JKF5 A0A2P8XP77 A0A2H8TJ53 A0A182MMH0 A0A182R9Q0 A0A1S4FFC6 A0A084VCS6 A0A182YFE8 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182JCK0 A0A2R7WM19 B0W8V5 A0A1B6JLJ0 A0A182F5H1 A0A182NEW7 A0A1W4W402 B4HWU4 A0A0J9QZV0 Q9VKR3 A0A1J1IXA1 B3N546 B3MJU6 B4GSZ1 B4P0T3 A0A3B0K1L3 Q29KQ9 A0A0L0BZD9 B4JE61 B4MWN9 A0A2S2PDY1 A0A0M4EGE6 A0A1A9XZJ2 W5JCY0 A0A1A9WZW6 B4KKP2 A0A1I8NM26 A0A1B0B900 A0A1I8MSU9 A0A336LN77 A0A0K8V585 Q7PW37 W8C942 A0A0R3NU97 A0A2S2Q5X0 Q173Z3 W8BRU6 A0A0J9R0F0 A0A182PDD4 M9MRT1 A0A0A1XM29 B4LUI3 A0A182KFF2 B4Q9S5 T1GFZ4 A0A1A9Z7T9 A0A1B0FLL2 A0A1A9UMK0 A0A182WGZ1 A0A1B0CVI9 A0A0N1PHU7 A0A212FF39 A0A2A4IV85 A0A194Q079 A0A3S2M6M2 A0A1J1IWQ7 A0A1B6EGK5 A0A0C9R6H2 K7IRS5 A0A1S3DMV9 A0A232F8T8 A0A195D3J4 A0A158NQR1 A0A195BEA4
Pubmed
22118469
26354079
20566863
18362917
19820115
24845553
+ More
23537049 29403074 24438588 25244985 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 26108605 20920257 23761445 25315136 12364791 24495485 17510324 25830018 20075255 28648823 21347285
23537049 29403074 24438588 25244985 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 26108605 20920257 23761445 25315136 12364791 24495485 17510324 25830018 20075255 28648823 21347285
EMBL
AGBW02008689
OWR52769.1
KQ460883
KPJ11231.1
KQ459603
KPI92725.1
+ More
ODYU01004657 SOQ44773.1 NWSH01005317 PCG64293.1 ODYU01003106 SOQ41443.1 ABLF02013798 ABLF02013799 ABLF02030156 GFXV01003509 MBW15314.1 GGMR01012271 MBY24890.1 DS235494 EEB16075.1 GECU01018020 JAS89686.1 ACPB03007015 NEVH01006721 PNF37114.1 DS497725 EFA12966.1 KK852543 KDR21672.1 GEDC01014844 JAS22454.1 APGK01018087 APGK01018088 APGK01018089 APGK01018090 APGK01018091 APGK01018092 KB740070 ENN81718.1 PYGN01001603 PSN33800.1 GFXV01001513 MBW13318.1 AXCM01005154 ATLV01010847 KE524620 KFB35770.1 AXCN02000839 APCN01003737 KK855069 PTY20664.1 DS231860 EDS39334.1 GECU01007744 JAS99962.1 CH480818 EDW52489.1 CM002910 KMY89607.1 AE014134 BT072821 AAF52997.3 ACN62424.1 CVRI01000063 CRL04728.1 CH954177 EDV58991.1 CH902620 EDV32401.1 CH479189 EDW25500.1 CM000157 EDW87978.2 OUUW01000006 SPP81760.1 CH379061 EAL33115.2 JRES01001119 KNC25385.1 CH916368 EDW03581.1 CH963857 EDW76528.1 GGMR01014986 MBY27605.1 CP012523 ALC39767.1 ADMH02001866 ETN60750.1 CH933807 EDW12706.2 JXJN01010172 JXJN01010173 JXJN01010174 JXJN01010175 JXJN01010176 UFQT01000031 SSX18231.1 GDHF01018225 JAI34089.1 AAAB01008984 EAA14739.3 GAMC01002484 JAC04072.1 KRT04753.1 GGMS01003952 MBY73155.1 CH477415 EAT41377.1 GAMC01002485 JAC04071.1 KMY89606.1 ADV37040.1 GBXI01001928 JAD12364.1 CH940649 EDW64169.2 CM000361 EDX04592.1 CAQQ02088126 CAQQ02088127 CAQQ02088128 CAQQ02088129 CCAG010020502 AJWK01030623 KQ460636 KPJ13388.1 AGBW02008864 OWR52338.1 NWSH01006480 PCG63406.1 KQ459592 KPI96820.1 RSAL01000020 RVE52624.1 CVRI01000061 CRL03996.1 GEDC01000231 JAS37067.1 GBYB01003585 JAG73352.1 NNAY01000701 OXU26910.1 KQ976885 KYN07473.1 ADTU01023471 KQ976511 KYM82527.1
ODYU01004657 SOQ44773.1 NWSH01005317 PCG64293.1 ODYU01003106 SOQ41443.1 ABLF02013798 ABLF02013799 ABLF02030156 GFXV01003509 MBW15314.1 GGMR01012271 MBY24890.1 DS235494 EEB16075.1 GECU01018020 JAS89686.1 ACPB03007015 NEVH01006721 PNF37114.1 DS497725 EFA12966.1 KK852543 KDR21672.1 GEDC01014844 JAS22454.1 APGK01018087 APGK01018088 APGK01018089 APGK01018090 APGK01018091 APGK01018092 KB740070 ENN81718.1 PYGN01001603 PSN33800.1 GFXV01001513 MBW13318.1 AXCM01005154 ATLV01010847 KE524620 KFB35770.1 AXCN02000839 APCN01003737 KK855069 PTY20664.1 DS231860 EDS39334.1 GECU01007744 JAS99962.1 CH480818 EDW52489.1 CM002910 KMY89607.1 AE014134 BT072821 AAF52997.3 ACN62424.1 CVRI01000063 CRL04728.1 CH954177 EDV58991.1 CH902620 EDV32401.1 CH479189 EDW25500.1 CM000157 EDW87978.2 OUUW01000006 SPP81760.1 CH379061 EAL33115.2 JRES01001119 KNC25385.1 CH916368 EDW03581.1 CH963857 EDW76528.1 GGMR01014986 MBY27605.1 CP012523 ALC39767.1 ADMH02001866 ETN60750.1 CH933807 EDW12706.2 JXJN01010172 JXJN01010173 JXJN01010174 JXJN01010175 JXJN01010176 UFQT01000031 SSX18231.1 GDHF01018225 JAI34089.1 AAAB01008984 EAA14739.3 GAMC01002484 JAC04072.1 KRT04753.1 GGMS01003952 MBY73155.1 CH477415 EAT41377.1 GAMC01002485 JAC04071.1 KMY89606.1 ADV37040.1 GBXI01001928 JAD12364.1 CH940649 EDW64169.2 CM000361 EDX04592.1 CAQQ02088126 CAQQ02088127 CAQQ02088128 CAQQ02088129 CCAG010020502 AJWK01030623 KQ460636 KPJ13388.1 AGBW02008864 OWR52338.1 NWSH01006480 PCG63406.1 KQ459592 KPI96820.1 RSAL01000020 RVE52624.1 CVRI01000061 CRL03996.1 GEDC01000231 JAS37067.1 GBYB01003585 JAG73352.1 NNAY01000701 OXU26910.1 KQ976885 KYN07473.1 ADTU01023471 KQ976511 KYM82527.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000007819
UP000009046
+ More
UP000015103 UP000235965 UP000007266 UP000027135 UP000019118 UP000079169 UP000245037 UP000075883 UP000075900 UP000030765 UP000076408 UP000075886 UP000076407 UP000075840 UP000075880 UP000002320 UP000069272 UP000075884 UP000192221 UP000001292 UP000000803 UP000183832 UP000008711 UP000007801 UP000008744 UP000002282 UP000268350 UP000001819 UP000037069 UP000001070 UP000007798 UP000092553 UP000092443 UP000000673 UP000091820 UP000009192 UP000095300 UP000092460 UP000095301 UP000007062 UP000008820 UP000075885 UP000008792 UP000075881 UP000000304 UP000015102 UP000092445 UP000092444 UP000078200 UP000075920 UP000092461 UP000283053 UP000002358 UP000215335 UP000078542 UP000005205 UP000078540
UP000015103 UP000235965 UP000007266 UP000027135 UP000019118 UP000079169 UP000245037 UP000075883 UP000075900 UP000030765 UP000076408 UP000075886 UP000076407 UP000075840 UP000075880 UP000002320 UP000069272 UP000075884 UP000192221 UP000001292 UP000000803 UP000183832 UP000008711 UP000007801 UP000008744 UP000002282 UP000268350 UP000001819 UP000037069 UP000001070 UP000007798 UP000092553 UP000092443 UP000000673 UP000091820 UP000009192 UP000095300 UP000092460 UP000095301 UP000007062 UP000008820 UP000075885 UP000008792 UP000075881 UP000000304 UP000015102 UP000092445 UP000092444 UP000078200 UP000075920 UP000092461 UP000283053 UP000002358 UP000215335 UP000078542 UP000005205 UP000078540
PRIDE
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A212FGA9
A0A194R5T7
A0A194PNM6
A0A2H1VVD4
A0A2A4IYV3
A0A2H1VKU5
+ More
X1WKQ9 J9K7I3 A0A2H8TMC7 A0A2S2P627 E0VRR9 A0A1B6IRW9 T1H8M0 A0A2J7R8F3 D7EK20 A0A067RD55 A0A1B6D9T4 N6TUH5 A0A1S3DUB1 J9JKF5 A0A2P8XP77 A0A2H8TJ53 A0A182MMH0 A0A182R9Q0 A0A1S4FFC6 A0A084VCS6 A0A182YFE8 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182JCK0 A0A2R7WM19 B0W8V5 A0A1B6JLJ0 A0A182F5H1 A0A182NEW7 A0A1W4W402 B4HWU4 A0A0J9QZV0 Q9VKR3 A0A1J1IXA1 B3N546 B3MJU6 B4GSZ1 B4P0T3 A0A3B0K1L3 Q29KQ9 A0A0L0BZD9 B4JE61 B4MWN9 A0A2S2PDY1 A0A0M4EGE6 A0A1A9XZJ2 W5JCY0 A0A1A9WZW6 B4KKP2 A0A1I8NM26 A0A1B0B900 A0A1I8MSU9 A0A336LN77 A0A0K8V585 Q7PW37 W8C942 A0A0R3NU97 A0A2S2Q5X0 Q173Z3 W8BRU6 A0A0J9R0F0 A0A182PDD4 M9MRT1 A0A0A1XM29 B4LUI3 A0A182KFF2 B4Q9S5 T1GFZ4 A0A1A9Z7T9 A0A1B0FLL2 A0A1A9UMK0 A0A182WGZ1 A0A1B0CVI9 A0A0N1PHU7 A0A212FF39 A0A2A4IV85 A0A194Q079 A0A3S2M6M2 A0A1J1IWQ7 A0A1B6EGK5 A0A0C9R6H2 K7IRS5 A0A1S3DMV9 A0A232F8T8 A0A195D3J4 A0A158NQR1 A0A195BEA4
X1WKQ9 J9K7I3 A0A2H8TMC7 A0A2S2P627 E0VRR9 A0A1B6IRW9 T1H8M0 A0A2J7R8F3 D7EK20 A0A067RD55 A0A1B6D9T4 N6TUH5 A0A1S3DUB1 J9JKF5 A0A2P8XP77 A0A2H8TJ53 A0A182MMH0 A0A182R9Q0 A0A1S4FFC6 A0A084VCS6 A0A182YFE8 A0A182QMI5 A0A2C9H8S2 A0A2C9GS68 A0A182JCK0 A0A2R7WM19 B0W8V5 A0A1B6JLJ0 A0A182F5H1 A0A182NEW7 A0A1W4W402 B4HWU4 A0A0J9QZV0 Q9VKR3 A0A1J1IXA1 B3N546 B3MJU6 B4GSZ1 B4P0T3 A0A3B0K1L3 Q29KQ9 A0A0L0BZD9 B4JE61 B4MWN9 A0A2S2PDY1 A0A0M4EGE6 A0A1A9XZJ2 W5JCY0 A0A1A9WZW6 B4KKP2 A0A1I8NM26 A0A1B0B900 A0A1I8MSU9 A0A336LN77 A0A0K8V585 Q7PW37 W8C942 A0A0R3NU97 A0A2S2Q5X0 Q173Z3 W8BRU6 A0A0J9R0F0 A0A182PDD4 M9MRT1 A0A0A1XM29 B4LUI3 A0A182KFF2 B4Q9S5 T1GFZ4 A0A1A9Z7T9 A0A1B0FLL2 A0A1A9UMK0 A0A182WGZ1 A0A1B0CVI9 A0A0N1PHU7 A0A212FF39 A0A2A4IV85 A0A194Q079 A0A3S2M6M2 A0A1J1IWQ7 A0A1B6EGK5 A0A0C9R6H2 K7IRS5 A0A1S3DMV9 A0A232F8T8 A0A195D3J4 A0A158NQR1 A0A195BEA4
PDB
2OXE
E-value=4.0768e-07,
Score=124
Ontologies
GO
PANTHER
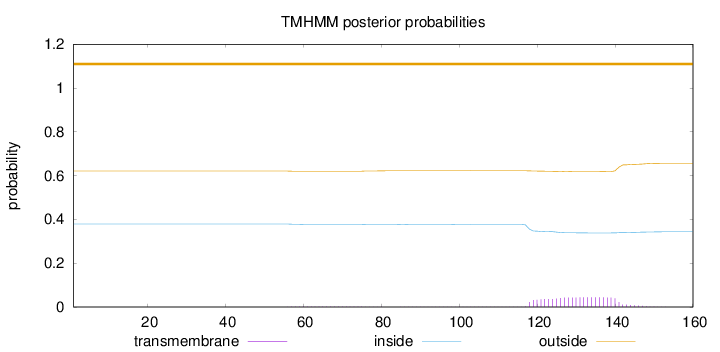
Topology
Subcellular location
Secreted
Length:
160
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.06607
Exp number, first 60 AAs:
0.00765
Total prob of N-in:
0.37931
outside
1 - 160
Population Genetic Test Statistics
Pi
274.814806
Theta
173.359423
Tajima's D
2.371906
CLR
0.073121
CSRT
0.932603369831508
Interpretation
Uncertain
