Gene
KWMTBOMO07141
Pre Gene Modal
BGIBMGA010551
Annotation
hypothetical_protein_KGM_22569_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.723 Nuclear Reliability : 2.226
Sequence
CDS
ATGGCAAATAATAGTTTGGAACATGATGATAATTGTTATAGTACGATAGATGAAAATATTTTTGATGATACATCTATAATTCCTTTAGAGAAGGATAGTTTCGCAATATGCGAATTGTGTGGTCGTGTTGGACGCCGAGGGCAGTTCTATCCAAGGAATAATAAATTTTGTTCACTTCGTTGCAACGCTAGTTTTTCGAGACGAAAACACATTAATTGGAGATTCAAACTAATGGAGGGTACAGATCACATGGCAGGGATGATTCCACTGGAGCCTTTACCCCAACTCCAGCAATGGCATGCCACTTTGAGTGATTTACAGGCAGGTCCCATTAAACAATCGGCAGCTTCAGTTGCGAACAGTTATGAATGGAAAGATGAGTTATTTGGTTGCGATTTCCTTGCTGCACCAGTGAGTCTTTTCAAACATGCACCTCTTCATGAAATGTGGGATAACACATTTGAGGGCATGAAGGTTGAAGTGAAGAATACAGACTGTGATAATTATTCTGAAAAACTTAGTGATTACTTTTGGGTAGCTACAGTTTTAAAAGTGAAAGGCTACATGGGACTTCTGAGATATGAAGGATTTGGGAGTGATGACTCCAAAGATTTTTGGGTGAATCTGTGCTGTTCAGAAGTTCATCCAGTTGGCTGGTGTGCTACACGAGGCAAACCACTCATACCACCACGAACTATAGAAGATAAATATACTGACTGGAAGAAGTTTCTTGTCAAACAACTAACGGGTGCACGTACACTACCGGCAAATTTTTATAGTAAACTGAACGACAGTCTTGTTTCTCGGTTTTCTATTGGATCATTAATGGAAGTTGTTGATAAAAATCGAATCTCACAAGTTAAGGTTGCTACAGTATGTGAAATAGTCGGCAAAAGATTGCACGTCAAATATTATGACAGCTCTCCGGAGGACAATGGTTTTTGGTGCCATGAAGATTCACCTCTTATACATCCAGTGGGCTGGGCGTTCCGGATCGGACATCCGTTAGATGCCCCTCAAAATTACTGTGAACGAGTTGCAACCGGTCGCTTATTTTCAAACGACAGTACAAATGACATGTTCTACAAATATCCTTCTAATGAACCACCATTGTTTATGGAGGGTATGAAAATCGAAGCCATAGACCCTTTGAATTTGTCAGCAGTATGTGCTGCTACAGTCATGCAGATATTGAATGAGGGTTATATGATGATAAGAATAGATTGCTATCCGGCTGATGCAAATGGCGCTGACTGGTTTTGCTACCACCAGAGATCGCCCTGTATTTTTCCGGTGGGGTTCGCTATGGCAAACAGCATTCCATTGGTTCCACCTGCAGGATTTTTGCCTGAGGAATTTCAATGGGAAGATTACTTGGCTATGACCGGAAGCATCGCGGCGGACCGATCCCTGTTCAGTGCTAGAGGTCACGTCGTTTCGCACGGCTTCGTCGCTGGAATGCGACTAGAATGTGCCGATCTTATGGACCCGAGGCTCGTTTGTGTCGCGACGATCGCTAGAGTTGTCGCGGACTTACTGAAGGTCCACTTCGACGGTTGGGGTTCGGAGTATGATCAGTGGTTATGGGCGCACAGCACCGACGTGTACCCGGTGGGATGGTGTCGCGCCATCGGACACCGGCTCGAGGGACCCCTGCAGCCGCCGCCACGGGTGCCCAGGAAGAACCAACCTCGGCAGCCCAAGAAGAAACGGAGACAAGTTACTAAAGTAGCCGCCCACACACTGAATTCTACAGAGGAGAGCTCCCAAAACTCAGACATCGGCGAAACAAAGAGTCTATCGCCGCAGACCAGCGTTTCTGAAGTATCAATGGACGCTGAAGCGGTTGTACCAAAAGACGAGCAATTGATTAGTGAAGTTAATATGGAATTACAACAAGAGCAAAAAGAAAAAACACCCGAGAAACCTAATGTATCTCCAATTTCGCCCAGGCATGATTTTATGGAGGACATTCCAATGCAGGACGAGAAACCTTCGATACCTGAAGAATCCCCTTCCAAATCTTCAGAAGGGTCGAGCAAAGAAGAAGAAGAGGATCCAGATAAAGTGATACCGAGACTAGTCAACACTACAGTGCCCTTAGATATATTAAATTCAGTTGATCCTGAAAACTGGACCGCGGCTGATGTTGCGAAATTTCTGACGGTAAACGACTGTCAGACGTACTGCGTTAATTTTTGCAATATAACAGGTCCGATGCTACTGCAGCTGTCTAAAGATGAAATCATAGAACTATTAGAAATGAAAGTTGGACCTTCACTTAAGATATTCGATTTAATACAACAACTGAAATGTAAAATTAAACAACCCCAGTGCAGAATGTTAGTAAATTTTAAGTAA
Protein
MANNSLEHDDNCYSTIDENIFDDTSIIPLEKDSFAICELCGRVGRRGQFYPRNNKFCSLRCNASFSRRKHINWRFKLMEGTDHMAGMIPLEPLPQLQQWHATLSDLQAGPIKQSAASVANSYEWKDELFGCDFLAAPVSLFKHAPLHEMWDNTFEGMKVEVKNTDCDNYSEKLSDYFWVATVLKVKGYMGLLRYEGFGSDDSKDFWVNLCCSEVHPVGWCATRGKPLIPPRTIEDKYTDWKKFLVKQLTGARTLPANFYSKLNDSLVSRFSIGSLMEVVDKNRISQVKVATVCEIVGKRLHVKYYDSSPEDNGFWCHEDSPLIHPVGWAFRIGHPLDAPQNYCERVATGRLFSNDSTNDMFYKYPSNEPPLFMEGMKIEAIDPLNLSAVCAATVMQILNEGYMMIRIDCYPADANGADWFCYHQRSPCIFPVGFAMANSIPLVPPAGFLPEEFQWEDYLAMTGSIAADRSLFSARGHVVSHGFVAGMRLECADLMDPRLVCVATIARVVADLLKVHFDGWGSEYDQWLWAHSTDVYPVGWCRAIGHRLEGPLQPPPRVPRKNQPRQPKKKRRQVTKVAAHTLNSTEESSQNSDIGETKSLSPQTSVSEVSMDAEAVVPKDEQLISEVNMELQQEQKEKTPEKPNVSPISPRHDFMEDIPMQDEKPSIPEESPSKSSEGSSKEEEEDPDKVIPRLVNTTVPLDILNSVDPENWTAADVAKFLTVNDCQTYCVNFCNITGPMLLQLSKDEIIELLEMKVGPSLKIFDLIQQLKCKIKQPQCRMLVNFK
Summary
Uniprot
H9JLZ9
A0A2A4JV94
A0A2H1VKT3
A0A212FGB3
A0A194R144
A0A2A4JVY7
+ More
A0A194PHD6 A0A067RMH1 F4WHY3 A0A195BEV1 A0A158NBX4 A0A0N0BIA0 A0A151WVF0 A0A0L7RK97 A0A0J7KYT1 A0A195FL65 A0A151IYY8 A0A2A3EH81 A0A232EVN0 A0A0T6BBA9 E9J683 A0A310S6L2 A0A154PPU8 A0A026X101 A0A3L8DSI4 A0A0C9R091 A0A0C9Q5Y2 E2A1V0 A0A1Y1KPA0 A0A088AC44 A0A1Y1KS40 E2BGP9 E0W3A5 A0A1S4N2J9 A0A336M8F5 A0A336KNL0 A0A336KPI6 N6UMN4 T1IEK5 A0A182H0J8 A0A1B6JUG9 A0A023F133 A0A0P5IRV2
A0A194PHD6 A0A067RMH1 F4WHY3 A0A195BEV1 A0A158NBX4 A0A0N0BIA0 A0A151WVF0 A0A0L7RK97 A0A0J7KYT1 A0A195FL65 A0A151IYY8 A0A2A3EH81 A0A232EVN0 A0A0T6BBA9 E9J683 A0A310S6L2 A0A154PPU8 A0A026X101 A0A3L8DSI4 A0A0C9R091 A0A0C9Q5Y2 E2A1V0 A0A1Y1KPA0 A0A088AC44 A0A1Y1KS40 E2BGP9 E0W3A5 A0A1S4N2J9 A0A336M8F5 A0A336KNL0 A0A336KPI6 N6UMN4 T1IEK5 A0A182H0J8 A0A1B6JUG9 A0A023F133 A0A0P5IRV2
Pubmed
EMBL
BABH01005568
NWSH01000511
PCG75945.1
ODYU01003105
SOQ41440.1
AGBW02008689
+ More
OWR52772.1 KQ460883 KPJ11234.1 PCG75946.1 KQ459603 KPI92722.1 KK852424 KDR24213.1 GL888170 EGI66212.1 KQ976500 KYM83103.1 ADTU01011455 KQ435735 KOX77141.1 KQ982702 KYQ51882.1 KQ414574 KOC71259.1 LBMM01001819 KMQ95687.1 KQ981491 KYN41007.1 KQ980732 KYN13962.1 KZ288258 PBC30516.1 NNAY01001980 OXU22389.1 LJIG01002339 KRT84601.1 GL768230 EFZ11648.1 KQ767687 OAD53354.1 KQ435017 KZC13892.1 KK107036 EZA61967.1 QOIP01000004 RLU23272.1 GBYB01009529 JAG79296.1 GBYB01009528 JAG79295.1 GL435806 EFN72589.1 GEZM01077626 JAV63232.1 GEZM01077627 JAV63231.1 GL448204 EFN85119.1 DS235882 EEB20111.1 AAZO01007355 UFQS01000603 UFQT01000603 SSX05280.1 SSX25641.1 UFQS01000665 UFQT01000665 SSX05947.1 SSX26306.1 SSX05948.1 SSX26307.1 APGK01008049 APGK01026394 KB740629 KB737568 ENN79936.1 ENN82995.1 ACPB03009805 JXUM01101441 JXUM01101442 KQ564649 KXJ71961.1 GECU01004894 JAT02813.1 GBBI01003485 JAC15227.1 GDIQ01211394 JAK40331.1
OWR52772.1 KQ460883 KPJ11234.1 PCG75946.1 KQ459603 KPI92722.1 KK852424 KDR24213.1 GL888170 EGI66212.1 KQ976500 KYM83103.1 ADTU01011455 KQ435735 KOX77141.1 KQ982702 KYQ51882.1 KQ414574 KOC71259.1 LBMM01001819 KMQ95687.1 KQ981491 KYN41007.1 KQ980732 KYN13962.1 KZ288258 PBC30516.1 NNAY01001980 OXU22389.1 LJIG01002339 KRT84601.1 GL768230 EFZ11648.1 KQ767687 OAD53354.1 KQ435017 KZC13892.1 KK107036 EZA61967.1 QOIP01000004 RLU23272.1 GBYB01009529 JAG79296.1 GBYB01009528 JAG79295.1 GL435806 EFN72589.1 GEZM01077626 JAV63232.1 GEZM01077627 JAV63231.1 GL448204 EFN85119.1 DS235882 EEB20111.1 AAZO01007355 UFQS01000603 UFQT01000603 SSX05280.1 SSX25641.1 UFQS01000665 UFQT01000665 SSX05947.1 SSX26306.1 SSX05948.1 SSX26307.1 APGK01008049 APGK01026394 KB740629 KB737568 ENN79936.1 ENN82995.1 ACPB03009805 JXUM01101441 JXUM01101442 KQ564649 KXJ71961.1 GECU01004894 JAT02813.1 GBBI01003485 JAC15227.1 GDIQ01211394 JAK40331.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000027135
+ More
UP000007755 UP000078540 UP000005205 UP000053105 UP000075809 UP000053825 UP000036403 UP000078541 UP000078492 UP000242457 UP000215335 UP000076502 UP000053097 UP000279307 UP000000311 UP000005203 UP000008237 UP000009046 UP000019118 UP000015103 UP000069940 UP000249989
UP000007755 UP000078540 UP000005205 UP000053105 UP000075809 UP000053825 UP000036403 UP000078541 UP000078492 UP000242457 UP000215335 UP000076502 UP000053097 UP000279307 UP000000311 UP000005203 UP000008237 UP000009046 UP000019118 UP000015103 UP000069940 UP000249989
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JLZ9
A0A2A4JV94
A0A2H1VKT3
A0A212FGB3
A0A194R144
A0A2A4JVY7
+ More
A0A194PHD6 A0A067RMH1 F4WHY3 A0A195BEV1 A0A158NBX4 A0A0N0BIA0 A0A151WVF0 A0A0L7RK97 A0A0J7KYT1 A0A195FL65 A0A151IYY8 A0A2A3EH81 A0A232EVN0 A0A0T6BBA9 E9J683 A0A310S6L2 A0A154PPU8 A0A026X101 A0A3L8DSI4 A0A0C9R091 A0A0C9Q5Y2 E2A1V0 A0A1Y1KPA0 A0A088AC44 A0A1Y1KS40 E2BGP9 E0W3A5 A0A1S4N2J9 A0A336M8F5 A0A336KNL0 A0A336KPI6 N6UMN4 T1IEK5 A0A182H0J8 A0A1B6JUG9 A0A023F133 A0A0P5IRV2
A0A194PHD6 A0A067RMH1 F4WHY3 A0A195BEV1 A0A158NBX4 A0A0N0BIA0 A0A151WVF0 A0A0L7RK97 A0A0J7KYT1 A0A195FL65 A0A151IYY8 A0A2A3EH81 A0A232EVN0 A0A0T6BBA9 E9J683 A0A310S6L2 A0A154PPU8 A0A026X101 A0A3L8DSI4 A0A0C9R091 A0A0C9Q5Y2 E2A1V0 A0A1Y1KPA0 A0A088AC44 A0A1Y1KS40 E2BGP9 E0W3A5 A0A1S4N2J9 A0A336M8F5 A0A336KNL0 A0A336KPI6 N6UMN4 T1IEK5 A0A182H0J8 A0A1B6JUG9 A0A023F133 A0A0P5IRV2
PDB
4C5H
E-value=2.7907e-157,
Score=1427
Ontologies
GO
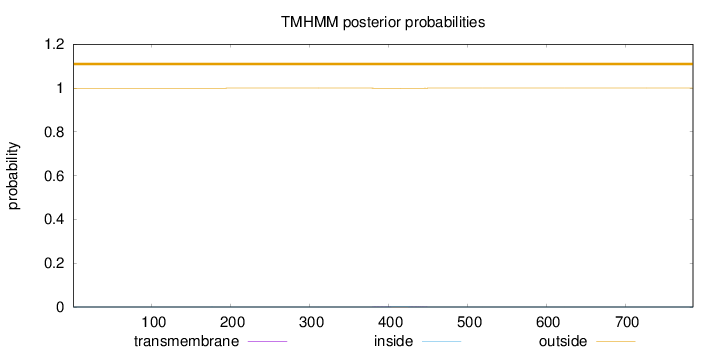
Topology
Length:
786
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02134
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00046
outside
1 - 786
Population Genetic Test Statistics
Pi
164.501048
Theta
147.383964
Tajima's D
0.625028
CLR
0.154497
CSRT
0.553722313884306
Interpretation
Uncertain
