Gene
KWMTBOMO07132 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010546
Annotation
prophenol_oxidase_activating_enzyme_precursor_[Manduca_sexta]
Location in the cell
Extracellular Reliability : 4.475
Sequence
CDS
ATGAGGAATAAAGTGTTGTTGCTTGTGTCGTCGCTTTTTTGGAGCTATGTTCTTTCAGATCATTGCACGACGCCGCTCGGCGAAGCTAGCGAATGCGTCTCATTGTACGATTGCTCTCAACTGTTGTCTGCGTTCGAGCAACGGCCCTTACAAAGCAAAGTGGTCAGCTTCCTGAGACAATCACAATGTGGTTTTCAGGGATACGTACCGAGAGTTTGTTGTGGGCCTTTACCGGCGCAACAAGAGAGGCAGACTACGCCACGCTCAAGAACAACGCGGCCTCCGACGAATGTGGTCGGCCACGTCGACCCCACAGTGCCAGAAGACTCGTCCCCAGCTCCTCGGAATCAATGCGGAGTGGACACACAAGGTGACAGAATCTATGGGGGTCAGTTCACGGACTTAGATGAATTTCCTTGGATGGCTTTGCTTGGATATCTGACGAGTAGGAATACCATAACCTATCAATGCGGCGGTGTGCTCTTCAACGCCCGCTACGTGCTAACGGCTGCTCATTGTTTGATCGGTGCAATCGAAAAGGAAGTCGGTAAATTATAA
Protein
MRNKVLLLVSSLFWSYVLSDHCTTPLGEASECVSLYDCSQLLSAFEQRPLQSKVVSFLRQSQCGFQGYVPRVCCGPLPAQQERQTTPRSRTTRPPTNVVGHVDPTVPEDSSPAPRNQCGVDTQGDRIYGGQFTDLDEFPWMALLGYLTSRNTITYQCGGVLFNARYVLTAAHCLIGAIEKEVGKL
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JLZ4
O77102
Q5DI99
A0A290U615
D5LPT4
I4DJ03
+ More
I4DN03 B3F5B1 A0A2A4JQA1 Q49QW1 A0A173GPF2 A0A0U1WT79 A0A212EKT5 A0A2H1V1Q2 A0A3S2M731 A0A194PP07 S4PK70 Q0VJV8 A0A194R0R2 V5G713 A0A0L7LLI6 F5HKX0 F4YJV1 A0A182HUC7 B1B5K1 J3JYT9 A0A154P5I7 N6TWW7 T1DTQ9 A0A2C9GLQ9 U4U606 A0A1S4HBZ1 A0A023EQB0 J3JXM3 W5JAY3 A0A182V2E3 A0A182LC02 A0A1W7R8M0 A0A336LMT0 A0A182TDG9 A0A182N6J8 A0A0K8VA86 A0A1W4XI92 U4USQ8 A0A182FAQ9 A0A182JX59 A0A0M9A7E2 A0A0K8WFT7 A0A182QUT5 A0A034V075 A0A0K8VB28 A0A067QHJ6 A0A182RLA4 A0A1Q3FC89 A0A0A1WGL4 A0A182XVS0 B0WA88 A0A182LWY5 A0A1J1I303 B4N8B1 A0A182ISD4 A0A084VDB9 A0A1Q3FEM6 A0A1B0FQW4 A0A1A9VW86 A0A1B0BD68 Q17EX4 A0A1A9X740 A0A1A9Z167 B0XAK3 A0A1Y1LC92 A0A154P3E5 A0A1Q3FC43 A0A232EZG2 A0A2J7Q299 Q6BD14 W8BLL3 A0A0C9QZC0 A0A0L7QXF0 B4M4J5 A0A2J7Q278 A0A1A9W879 A0A2P8YP04 A0A1Y1LA48 A0A1Y1LA41 A0A1V1G586 E0VXK9 A0A2J7Q286 B4QZQ4 K7J980 B4I4W4 B4PRF7 A0A182JMK2 Q6Y2X4 Q5DI98 A0A310SR39 A0A2M4AVL9 A0A182QL12 A0A1L8DQN9
I4DN03 B3F5B1 A0A2A4JQA1 Q49QW1 A0A173GPF2 A0A0U1WT79 A0A212EKT5 A0A2H1V1Q2 A0A3S2M731 A0A194PP07 S4PK70 Q0VJV8 A0A194R0R2 V5G713 A0A0L7LLI6 F5HKX0 F4YJV1 A0A182HUC7 B1B5K1 J3JYT9 A0A154P5I7 N6TWW7 T1DTQ9 A0A2C9GLQ9 U4U606 A0A1S4HBZ1 A0A023EQB0 J3JXM3 W5JAY3 A0A182V2E3 A0A182LC02 A0A1W7R8M0 A0A336LMT0 A0A182TDG9 A0A182N6J8 A0A0K8VA86 A0A1W4XI92 U4USQ8 A0A182FAQ9 A0A182JX59 A0A0M9A7E2 A0A0K8WFT7 A0A182QUT5 A0A034V075 A0A0K8VB28 A0A067QHJ6 A0A182RLA4 A0A1Q3FC89 A0A0A1WGL4 A0A182XVS0 B0WA88 A0A182LWY5 A0A1J1I303 B4N8B1 A0A182ISD4 A0A084VDB9 A0A1Q3FEM6 A0A1B0FQW4 A0A1A9VW86 A0A1B0BD68 Q17EX4 A0A1A9X740 A0A1A9Z167 B0XAK3 A0A1Y1LC92 A0A154P3E5 A0A1Q3FC43 A0A232EZG2 A0A2J7Q299 Q6BD14 W8BLL3 A0A0C9QZC0 A0A0L7QXF0 B4M4J5 A0A2J7Q278 A0A1A9W879 A0A2P8YP04 A0A1Y1LA48 A0A1Y1LA41 A0A1V1G586 E0VXK9 A0A2J7Q286 B4QZQ4 K7J980 B4I4W4 B4PRF7 A0A182JMK2 Q6Y2X4 Q5DI98 A0A310SR39 A0A2M4AVL9 A0A182QL12 A0A1L8DQN9
Pubmed
19121390
9770467
15857768
18207075
28953952
22651552
+ More
19557749 22118469 26354079 23622113 26227816 12364791 14747013 17210077 18195005 22516182 23537049 24945155 20920257 23761445 20966253 25348373 24845553 25830018 25244985 17994087 24438588 17510324 28004739 28648823 15579698 24495485 29403074 28410430 20566863 20075255 17550304 14505699 16033436
19557749 22118469 26354079 23622113 26227816 12364791 14747013 17210077 18195005 22516182 23537049 24945155 20920257 23761445 20966253 25348373 24845553 25830018 25244985 17994087 24438588 17510324 28004739 28648823 15579698 24495485 29403074 28410430 20566863 20075255 17550304 14505699 16033436
EMBL
BABH01005537
BABH01005538
AF059728
AAC64004.1
AY789465
AM293327
+ More
AAX18636.1 CAL25132.1 KY680243 ATD13324.1 GU953224 ADF43208.1 AK401271 BAM17893.1 AK402671 BAM19293.1 EF614332 ABU98654.1 NWSH01000833 PCG73946.1 AY677081 AAW24480.1 KT006136 ANH58161.1 JQ581597 AFX82593.1 AGBW02014220 OWR42068.1 ODYU01000283 SOQ34775.1 RSAL01000017 RVE52975.1 KQ459603 KPI92860.1 GAIX01004665 JAA87895.1 AM293316 CAL25121.1 KQ460883 KPJ11099.1 GALX01002586 JAB65880.1 JTDY01000666 KOB76290.1 AAAB01008879 EGK96931.1 HM070255 ADZ72972.1 APCN01000619 AB363980 BAG14262.2 BT128420 AEE63377.1 KQ434809 KZC06390.1 APGK01057809 KB741282 ENN70807.1 GAMD01000809 JAB00782.1 KB631684 ERL85365.1 GAPW01001836 JAC11762.1 BT127992 AEE62954.1 ADMH02001903 ETN60568.1 GEHC01000168 JAV47477.1 UFQT01000091 SSX19534.1 GDHF01016430 JAI35884.1 KB632345 ERL93135.1 KQ435719 KOX78755.1 GDHF01002415 JAI49899.1 AXCN02000286 GAKP01023416 JAC35542.1 GDHF01016160 JAI36154.1 KK853628 KDR03824.1 GFDL01009851 JAV25194.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1 DS231869 EDS40917.1 AXCM01000870 CVRI01000039 CRK94579.1 CH964232 EDW81362.1 ATLV01011243 ATLV01011244 ATLV01011245 ATLV01011246 KE524657 KFB35963.1 GFDL01009047 JAV25998.1 CCAG010023843 JXJN01012354 CH477278 EAT45085.1 DS232586 EDS43739.1 GEZM01061361 GEZM01061358 GEZM01061351 JAV70518.1 KZC06391.1 GFDL01009899 JAV25146.1 NNAY01001477 OXU23832.1 NEVH01019373 PNF22708.1 AY665377 AAT76544.1 GAMC01016016 JAB90539.1 GBYB01001065 JAG70832.1 KQ414704 KOC63285.1 CH940652 EDW59556.1 PNF22686.1 PYGN01000457 PSN45975.1 GEZM01061360 JAV70519.1 GEZM01061363 JAV70514.1 FX985343 BAX07356.1 DS235833 EEB18115.1 PNF22687.1 CM000364 EDX12012.1 AAZX01002213 CH480821 EDW55257.1 CM000160 EDW96345.1 AY188445 AAO74570.1 AH014670 AY789466 AAX18637.1 KQ761039 OAD58341.1 GGFK01011500 MBW44821.1 AXCN02000165 GFDF01005424 JAV08660.1
AAX18636.1 CAL25132.1 KY680243 ATD13324.1 GU953224 ADF43208.1 AK401271 BAM17893.1 AK402671 BAM19293.1 EF614332 ABU98654.1 NWSH01000833 PCG73946.1 AY677081 AAW24480.1 KT006136 ANH58161.1 JQ581597 AFX82593.1 AGBW02014220 OWR42068.1 ODYU01000283 SOQ34775.1 RSAL01000017 RVE52975.1 KQ459603 KPI92860.1 GAIX01004665 JAA87895.1 AM293316 CAL25121.1 KQ460883 KPJ11099.1 GALX01002586 JAB65880.1 JTDY01000666 KOB76290.1 AAAB01008879 EGK96931.1 HM070255 ADZ72972.1 APCN01000619 AB363980 BAG14262.2 BT128420 AEE63377.1 KQ434809 KZC06390.1 APGK01057809 KB741282 ENN70807.1 GAMD01000809 JAB00782.1 KB631684 ERL85365.1 GAPW01001836 JAC11762.1 BT127992 AEE62954.1 ADMH02001903 ETN60568.1 GEHC01000168 JAV47477.1 UFQT01000091 SSX19534.1 GDHF01016430 JAI35884.1 KB632345 ERL93135.1 KQ435719 KOX78755.1 GDHF01002415 JAI49899.1 AXCN02000286 GAKP01023416 JAC35542.1 GDHF01016160 JAI36154.1 KK853628 KDR03824.1 GFDL01009851 JAV25194.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1 DS231869 EDS40917.1 AXCM01000870 CVRI01000039 CRK94579.1 CH964232 EDW81362.1 ATLV01011243 ATLV01011244 ATLV01011245 ATLV01011246 KE524657 KFB35963.1 GFDL01009047 JAV25998.1 CCAG010023843 JXJN01012354 CH477278 EAT45085.1 DS232586 EDS43739.1 GEZM01061361 GEZM01061358 GEZM01061351 JAV70518.1 KZC06391.1 GFDL01009899 JAV25146.1 NNAY01001477 OXU23832.1 NEVH01019373 PNF22708.1 AY665377 AAT76544.1 GAMC01016016 JAB90539.1 GBYB01001065 JAG70832.1 KQ414704 KOC63285.1 CH940652 EDW59556.1 PNF22686.1 PYGN01000457 PSN45975.1 GEZM01061360 JAV70519.1 GEZM01061363 JAV70514.1 FX985343 BAX07356.1 DS235833 EEB18115.1 PNF22687.1 CM000364 EDX12012.1 AAZX01002213 CH480821 EDW55257.1 CM000160 EDW96345.1 AY188445 AAO74570.1 AH014670 AY789466 AAX18637.1 KQ761039 OAD58341.1 GGFK01011500 MBW44821.1 AXCN02000165 GFDF01005424 JAV08660.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000037510 UP000007062 UP000075840 UP000076502 UP000019118 UP000030742 UP000000673 UP000075903 UP000075882 UP000075902 UP000075884 UP000192223 UP000069272 UP000075881 UP000053105 UP000075886 UP000027135 UP000075900 UP000076408 UP000002320 UP000075883 UP000183832 UP000007798 UP000075880 UP000030765 UP000092444 UP000078200 UP000092460 UP000008820 UP000092443 UP000092445 UP000215335 UP000235965 UP000053825 UP000008792 UP000091820 UP000245037 UP000009046 UP000000304 UP000002358 UP000001292 UP000002282
UP000037510 UP000007062 UP000075840 UP000076502 UP000019118 UP000030742 UP000000673 UP000075903 UP000075882 UP000075902 UP000075884 UP000192223 UP000069272 UP000075881 UP000053105 UP000075886 UP000027135 UP000075900 UP000076408 UP000002320 UP000075883 UP000183832 UP000007798 UP000075880 UP000030765 UP000092444 UP000078200 UP000092460 UP000008820 UP000092443 UP000092445 UP000215335 UP000235965 UP000053825 UP000008792 UP000091820 UP000245037 UP000009046 UP000000304 UP000002358 UP000001292 UP000002282
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JLZ4
O77102
Q5DI99
A0A290U615
D5LPT4
I4DJ03
+ More
I4DN03 B3F5B1 A0A2A4JQA1 Q49QW1 A0A173GPF2 A0A0U1WT79 A0A212EKT5 A0A2H1V1Q2 A0A3S2M731 A0A194PP07 S4PK70 Q0VJV8 A0A194R0R2 V5G713 A0A0L7LLI6 F5HKX0 F4YJV1 A0A182HUC7 B1B5K1 J3JYT9 A0A154P5I7 N6TWW7 T1DTQ9 A0A2C9GLQ9 U4U606 A0A1S4HBZ1 A0A023EQB0 J3JXM3 W5JAY3 A0A182V2E3 A0A182LC02 A0A1W7R8M0 A0A336LMT0 A0A182TDG9 A0A182N6J8 A0A0K8VA86 A0A1W4XI92 U4USQ8 A0A182FAQ9 A0A182JX59 A0A0M9A7E2 A0A0K8WFT7 A0A182QUT5 A0A034V075 A0A0K8VB28 A0A067QHJ6 A0A182RLA4 A0A1Q3FC89 A0A0A1WGL4 A0A182XVS0 B0WA88 A0A182LWY5 A0A1J1I303 B4N8B1 A0A182ISD4 A0A084VDB9 A0A1Q3FEM6 A0A1B0FQW4 A0A1A9VW86 A0A1B0BD68 Q17EX4 A0A1A9X740 A0A1A9Z167 B0XAK3 A0A1Y1LC92 A0A154P3E5 A0A1Q3FC43 A0A232EZG2 A0A2J7Q299 Q6BD14 W8BLL3 A0A0C9QZC0 A0A0L7QXF0 B4M4J5 A0A2J7Q278 A0A1A9W879 A0A2P8YP04 A0A1Y1LA48 A0A1Y1LA41 A0A1V1G586 E0VXK9 A0A2J7Q286 B4QZQ4 K7J980 B4I4W4 B4PRF7 A0A182JMK2 Q6Y2X4 Q5DI98 A0A310SR39 A0A2M4AVL9 A0A182QL12 A0A1L8DQN9
I4DN03 B3F5B1 A0A2A4JQA1 Q49QW1 A0A173GPF2 A0A0U1WT79 A0A212EKT5 A0A2H1V1Q2 A0A3S2M731 A0A194PP07 S4PK70 Q0VJV8 A0A194R0R2 V5G713 A0A0L7LLI6 F5HKX0 F4YJV1 A0A182HUC7 B1B5K1 J3JYT9 A0A154P5I7 N6TWW7 T1DTQ9 A0A2C9GLQ9 U4U606 A0A1S4HBZ1 A0A023EQB0 J3JXM3 W5JAY3 A0A182V2E3 A0A182LC02 A0A1W7R8M0 A0A336LMT0 A0A182TDG9 A0A182N6J8 A0A0K8VA86 A0A1W4XI92 U4USQ8 A0A182FAQ9 A0A182JX59 A0A0M9A7E2 A0A0K8WFT7 A0A182QUT5 A0A034V075 A0A0K8VB28 A0A067QHJ6 A0A182RLA4 A0A1Q3FC89 A0A0A1WGL4 A0A182XVS0 B0WA88 A0A182LWY5 A0A1J1I303 B4N8B1 A0A182ISD4 A0A084VDB9 A0A1Q3FEM6 A0A1B0FQW4 A0A1A9VW86 A0A1B0BD68 Q17EX4 A0A1A9X740 A0A1A9Z167 B0XAK3 A0A1Y1LC92 A0A154P3E5 A0A1Q3FC43 A0A232EZG2 A0A2J7Q299 Q6BD14 W8BLL3 A0A0C9QZC0 A0A0L7QXF0 B4M4J5 A0A2J7Q278 A0A1A9W879 A0A2P8YP04 A0A1Y1LA48 A0A1Y1LA41 A0A1V1G586 E0VXK9 A0A2J7Q286 B4QZQ4 K7J980 B4I4W4 B4PRF7 A0A182JMK2 Q6Y2X4 Q5DI98 A0A310SR39 A0A2M4AVL9 A0A182QL12 A0A1L8DQN9
PDB
2OLG
E-value=3.88053e-13,
Score=177
Ontologies
GO
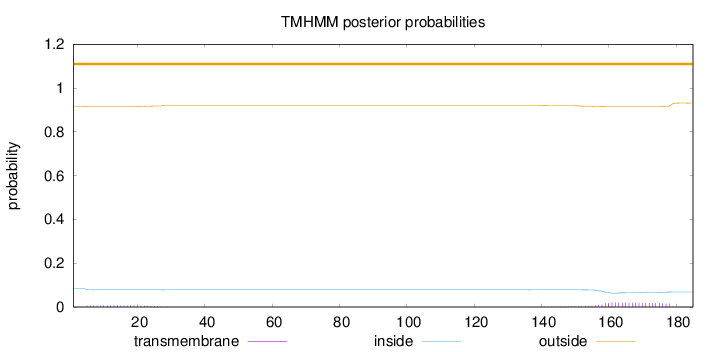
Topology
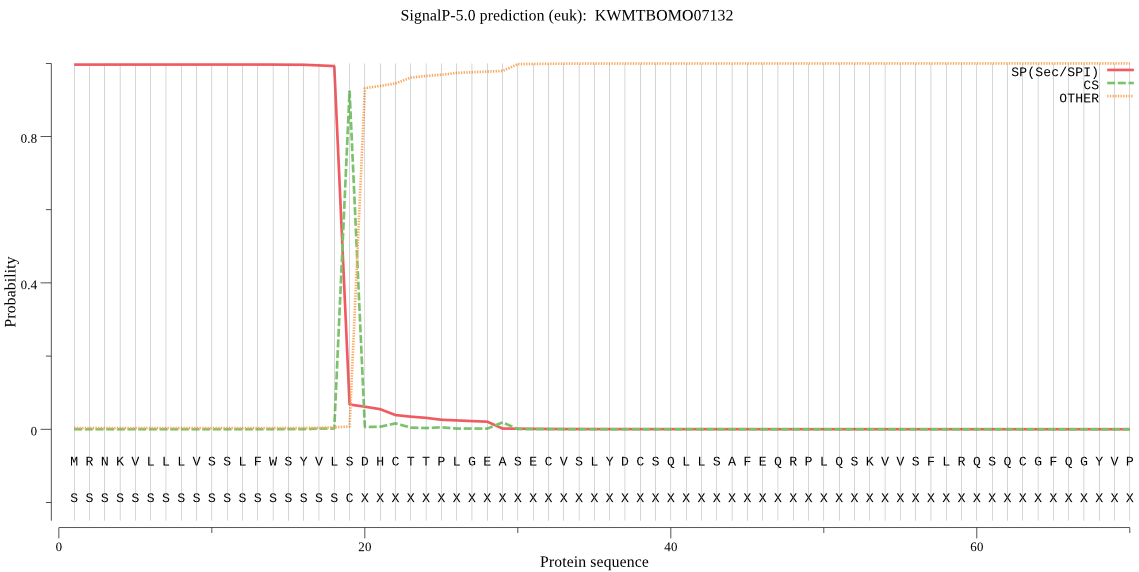
SignalP
Position: 1 - 19,
Likelihood: 0.996506
Length:
185
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.58247
Exp number, first 60 AAs:
0.1316
Total prob of N-in:
0.08462
outside
1 - 185
Population Genetic Test Statistics
Pi
193.427763
Theta
148.393119
Tajima's D
0.632672
CLR
0.659873
CSRT
0.55777211139443
Interpretation
Uncertain
