Gene
KWMTBOMO07126 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010409
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.17
Sequence
CDS
ATGGAAGACAATAGTGAAAGTGTTAGTGTCAGTGACAGTGATTTTGTCGGGTTCCTCCGGGAAAGGTACCCGACAATTAGGGCCGAGTATCTTAGGTATAGGGCCTCTCGTGGCAATCCCTCCCCCCCCGCGTCCGTCGCCGCGTTTCTCAACCGTGCCTCGGAGGCACGCCGCGCGCCCCCCGCCGCCGCGTTAAGTTCGAGCGCACGTTCCGACGATGCGGCTCACATAGCGCCTAACGCTACCCCCACCCCCGCGCCCGCGTCGTTTACGTCATCGCGCGCTTCGTCCGTCGCGACCTCGATTGTTTCGAGTTCAGGCTCCGATACCGAATCGGAGATGGATTTCGAATCCGCGTCAAGCCCTCAGCCTGGAACTTCGGATGGGTTCCAAACCGTAACGCGCGGTAAAAAGCGTACTCGCGCCGTGGAGTCCCGGAGCTCCACGACGAAGCAGACAAAATCCGCGACCGCCTCCCGCCCGCAGGTAGTCGTGACACCGGAGTCGGACTCCGCTCGCCGCGTAACCCCGCCGCCGCGTCCCAAGCCCGCGCCCGCTCCTAAAGCAGCTGCCCCGCCCCCGCTGATTCTCCAGGAAAAAACTGCGTGGAACCGCGTTTCCCAGGCCCTTCAGGCAAATAAAATTAATTATACCCATGCGCGTAACGTCGCGCATGGGATTCAGATAAAGGTCGCAACGCCGGGCGACCATAGGGCCCTCTCAGCATACCTCCGAAAGGAGAACATAGGTTTCCACACCTATGCTCTTCAGGAGGACCGCGAGCTCCGCGTAGTGATACGCGGAGTCCCTAAAGAACTCGACATCGACTACATAAAGGAGGATCTGACCGCTCAGGCCCTCCCGATAGTTAGTGTGCACCGGATGCACAGCGGGCGGGGCAAACAGCCCTATAATATGATACTCGTGGCGTTAGAACCCACCCCGGAGGCCAAAAAGAAGATCGCATGTCTCACGACAATTTGCGGCCTATCCGGGATCTCGATCGAAGCCCCCCATAAGCGTGGCACTCCCGGGCAGTGCTACAGGTGCCAACTCTATGGCCACTCAGCGCGTAATTGCCACGCGCGCCCCCGCTGCGTGAAATGCCTCGGCGATCACGCTACGTTAGATTGCGTTAGAGATAGGGAAACCGCGACCGAACCTCCCAGCTGTGTCCTATGCCTTAAGCAAGGGCACCCCGCGAACTACCGTGGATGTCCCAGGGCTCCGCGGAAACGTCCAACCCACCCAGCTCCGCGAACCGTCGGCCCAAAGACTTCGGCACCTTCAGTGCCGAAACCCGCCTTCGTGCCGGCTGCAGTTCCCACGGTCTCGGCGTGGAAAAAGCCGCTGCCGTATACGAAGGCGGGAACGGAAACCGCACCCCCGCCCCCGCTCGCGACACGCCCCGCGCCTCAGCCCCTGCTCGCACCTCGCCCCGCGCCCGCGTTCCGTCCCCCGCAACCCTCTGCCGCCAACGACTTCGCGCTCGTGCGCGACTTCGTCACCGCGGTGAACTTCGACCGTCTGCGATCGTTCGCTGACGCTATCCGTAGGTCGACAACCCCCGAACAGCGGCTCGCGGCCGCGTTCGATCACATGGACGTTTACGAGTCCGTGTCGCCATTCTATAATGCTAACGGACTCGCGCGGCAACGCGATCAAATTTTTGAATTCCTCCGCGATAATCTTGTAGATATTCTATTAGTGCAGGAAACCTGTCTGAAGCCCTCGCGTCGCGACCCGAAAGTCGCGAACTACGTCATGGTTAGGAATGACAGACTCACCGCCTCCAAAGGCGGGACTGCCATTTACTATAGGCGGGCCCTGCACGTTGTCCCTCTCGATACTCCCTCGCTCTCACATATCGAGGCGTCAGTGTGCCGTATCTCGCTGACGGGACACCAGCCGATCGTCATCGCATCCGTTTATCTCCCCCCGGACAAGCCCCTCCTGAGCAGTGACATCGAGTCACTGTTCGGCATGGGAGACTCTGTCATCCTGGCAGGCGATTTAAATTGCCACCACACTAGGTGGAACTGCCATCGTACAAACGTTAACGGTAGGCGTCTCGACGCGTTTATAGACGACCTCACCTTTGAAATAGTCGGTCCCCCAACTCCAACATGTTATCCGTATAACATCGCGCTCCGTCCGAGCACTATAGACCTGGCATTGCTTAGGAACGTAACTCTGCGCTTGCGTTCCATCGAAGCAATGTCAGAGCTCGACTCAGACCACCGACCTGTCGTTATGCAGCTCGGTCGCCCTCACAACCCAGTCACTGTTACGAGGACCATGGTGGATTGGAATAAGCTGGGCACGTGCCTAGCCGACGCCGCTCCGCCAATCCTCCCTTACGGCCCGGATTCGAATCCATCCCCCGAGGACACCGTCGAATCCATAAACATCATTACCGATCACATCTCTTCCGCGATCATTAGATCTTCTAAAGAAGTCGATGTGGAGGACAGCTTCCACCGCATCAGACTGTCCCCCGATCTTAGGAATCTCTTAAGAGTTAGGAACGCGGCAATCCGGGCCTACGATCGTCTTCCCACGCATTCAAACCGGATTCACATGCGTCGTCTACAACGCGAAGTCCACTCCCGCTTAAGCGACGCGCGTAACGATAATTGGCATAGTTATTTAGAACAACTCGCGCCCTCCCACCAAGCATACTGGCGACTAGCTAGGACTCTCAAATCCGAAACTACCGCTACTATGCCTCCCCTCTTACGCCCTTCAGGCCAACCACCGGCATTCGATGACGATGACAAGGCTGAGCTGCTGGCCGATGCACTGCAAGAGCAGTGCACCACCAGCACTCAACACGCGGACCCCGAACACACCGAGTTAGTCGACAGGGAGGTCGAGCGCAGAGCTTCCCTGCCGCCCTCGGACGCGTTACCCCCCATTACCACTGACGAAGTTAGAGACGCGATCCACAACCTCCAACCTAGGAAGGCACCAGGCTCCGACGGCATCCACAACCGCGCGCTAAAATTTTTGCCAGTCCAACTGATAGCAATGTTGGCTACAATCTTAAATGCCGCTATGACGCACTGCATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGTATACATAAGCCGGGCAAACCGAGAAACGAAACTTCTAGTTACCGTCCGATTAGTCTCCTCTCGACGATAGGAAAAATTTACGAACGTCTCCTTAGGAAACGCCTCTGGGATTTTGTTACCGCGAACAAAATTCTCATAGACGAACAGTTCGGATTCCGCTCAAAACACTCGTGCGTACAACAAGTGCACCGCCTCACGGAGCACATTCTGATAGGACTAAATAGGCGTAAGCAAATCCCGACCGGCGCCCTCTTCTTCGACATCGCGAAGGCGTTCGACAAAGTCTGGCACAACGGTTTGATCTACAAACTGTACAACATGGGAGTGCCAGACAGACTCGTGCTCATCATACGAGACTTCTTGTCGAACCGTTCGTTTCGATATCGAGTAGAGGGAACTCGTTCTCGTCCCCGTCAACTGACTGCCGGAGTCCCGCAAGGCTCCGCGCTCTCCCCGTTATTATTTTGTTTGTATATCAATGATATACCCCGGTCTCCGGAGACCCATCTAGCGCTCTTCGCCGACGACACGGCTATCTACTACTCGTGTAGGAAGATGTCGCTGCTTCATCGGCGACTCCAGATCGCAGTAGCCACCATGGGACAGTGGTTCCGGAAGTGGCGAATAGACATCAACCCCACGAAAAGCGCAGCGGTGCTCTTCAAAAGGGGTCGCCCTCCGAACATCACTTCGAGCATCCCACTCCGTAGTAGGCGCGCAAACACCTCCGCCGTTAGTCCCATCACTCTCTTTGGCCAGCCCATACCGTGGGTCTCGAAGGTCAAATATCTAGGCGTCACCCTCGACAGAGGGATGACATTCCGTCCCCATATAAAAACGGTTCGCGACCGCGCCGCGTTTATTCTAGGACGACTCTATCCAATGCTTTGTAGTCGAAGCAAACTGTCCCTCCGCAATAAGGTAACTCTCTACAAAACTTGTATACGCCCCGTCATGACGTATGCAAGCGTAGTGTTCGCTCACTCAGCCCGCACCCACTTGAAATCCCTTCAGGTTATTCAATCACGATTCTGCAGGATAGCCGTCGGAGCGCCATGGTTCCTTAGGAATGTGGATCTCCACGATGACCTGGAGCTCGACTCTCTTAAAACTAGTTTAGAAACCAGATTTCGCCTGCACAAAATGCCTGCCAGAAACAAGGACCAAGAACAAGAGGTTCTCACTTGGATCTCTGATGTTTTGGGAGAGCCGCTACCAAATGGCGCATACGAGGATGTGCTAAGGGACGGTGTGATCCTTTGCAAATTGGCCAACAAATTGGCTCCTGGATCAGTGAAAAAAATCCAAGAGAAAGGGACCAACTTCCAGCTCATGGAGAACATTCAAAGGTTCCAAGCGGCAATCAAGAAATACGGAGTACCAGAAGAAGAAATATTCCAAACCGCCGATCTTTTTGAGAGGCGTAACATTCCCCAAGTAACATTATGTTTGTACGCTCTGGGAAGAATTACTCAAAAACATCCCGAGTTTACAGGACCTCAGCTAGGACCTAAAATGGCTGATAAGAACGAACGTACCTTCACGGAAGAACAACTTCGGGCCCACAACGCTGAACTTAACCTCCAAATGGGCTTCAATAAGGGCGCCTCCCAGTCCGGTCATGGTGGCTTCGGTAACACCCGCCACATGTAA
Protein
MEDNSESVSVSDSDFVGFLRERYPTIRAEYLRYRASRGNPSPPASVAAFLNRASEARRAPPAAALSSSARSDDAAHIAPNATPTPAPASFTSSRASSVATSIVSSSGSDTESEMDFESASSPQPGTSDGFQTVTRGKKRTRAVESRSSTTKQTKSATASRPQVVVTPESDSARRVTPPPRPKPAPAPKAAAPPPLILQEKTAWNRVSQALQANKINYTHARNVAHGIQIKVATPGDHRALSAYLRKENIGFHTYALQEDRELRVVIRGVPKELDIDYIKEDLTAQALPIVSVHRMHSGRGKQPYNMILVALEPTPEAKKKIACLTTICGLSGISIEAPHKRGTPGQCYRCQLYGHSARNCHARPRCVKCLGDHATLDCVRDRETATEPPSCVLCLKQGHPANYRGCPRAPRKRPTHPAPRTVGPKTSAPSVPKPAFVPAAVPTVSAWKKPLPYTKAGTETAPPPPLATRPAPQPLLAPRPAPAFRPPQPSAANDFALVRDFVTAVNFDRLRSFADAIRRSTTPEQRLAAAFDHMDVYESVSPFYNANGLARQRDQIFEFLRDNLVDILLVQETCLKPSRRDPKVANYVMVRNDRLTASKGGTAIYYRRALHVVPLDTPSLSHIEASVCRISLTGHQPIVIASVYLPPDKPLLSSDIESLFGMGDSVILAGDLNCHHTRWNCHRTNVNGRRLDAFIDDLTFEIVGPPTPTCYPYNIALRPSTIDLALLRNVTLRLRSIEAMSELDSDHRPVVMQLGRPHNPVTVTRTMVDWNKLGTCLADAAPPILPYGPDSNPSPEDTVESINIITDHISSAIIRSSKEVDVEDSFHRIRLSPDLRNLLRVRNAAIRAYDRLPTHSNRIHMRRLQREVHSRLSDARNDNWHSYLEQLAPSHQAYWRLARTLKSETTATMPPLLRPSGQPPAFDDDDKAELLADALQEQCTTSTQHADPEHTELVDREVERRASLPPSDALPPITTDEVRDAIHNLQPRKAPGSDGIHNRALKFLPVQLIAMLATILNAAMTHCIFPAVWKEADVIGIHKPGKPRNETSSYRPISLLSTIGKIYERLLRKRLWDFVTANKILIDEQFGFRSKHSCVQQVHRLTEHILIGLNRRKQIPTGALFFDIAKAFDKVWHNGLIYKLYNMGVPDRLVLIIRDFLSNRSFRYRVEGTRSRPRQLTAGVPQGSALSPLLFCLYINDIPRSPETHLALFADDTAIYYSCRKMSLLHRRLQIAVATMGQWFRKWRIDINPTKSAAVLFKRGRPPNITSSIPLRSRRANTSAVSPITLFGQPIPWVSKVKYLGVTLDRGMTFRPHIKTVRDRAAFILGRLYPMLCSRSKLSLRNKVTLYKTCIRPVMTYASVVFAHSARTHLKSLQVIQSRFCRIAVGAPWFLRNVDLHDDLELDSLKTSLETRFRLHKMPARNKDQEQEVLTWISDVLGEPLPNGAYEDVLRDGVILCKLANKLAPGSVKKIQEKGTNFQLMENIQRFQAAIKKYGVPEEEIFQTADLFERRNIPQVTLCLYALGRITQKHPEFTGPQLGPKMADKNERTFTEEQLRAHNAELNLQMGFNKGASQSGHGGFGNTRHM
Summary
Uniprot
ProteinModelPortal
PDB
1WYP
E-value=2.59045e-18,
Score=232
Ontologies
GO
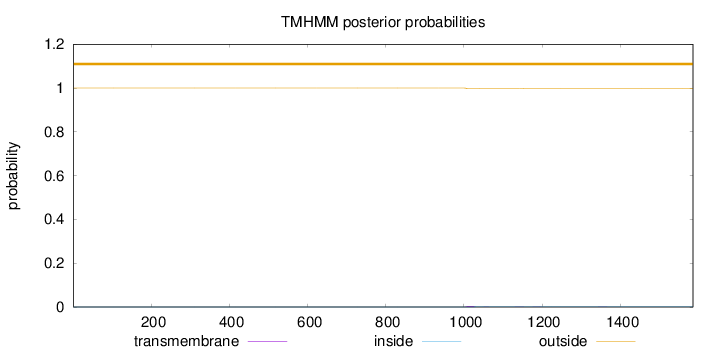
Topology
Length:
1586
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0966699999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1586
Population Genetic Test Statistics
Pi
174.403483
Theta
160.203312
Tajima's D
0.150934
CLR
0.090529
CSRT
0.407329633518324
Interpretation
Uncertain
