Gene
KWMTBOMO07114 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010419
Annotation
saposin-like_protein_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.737 Nuclear Reliability : 1.625
Sequence
CDS
ATGACAAACACGTTTGCTGTTTGTCTACTGTCTCTAACGTTCCTTTGTTGTACAAATTTGTCGTTTGCGCGACAAGTACCGAAGGAATGTGCTAAGGGACCACAAGTATGGTGCGAGAGTCTAAAGCGGGGGGCTGAATGCGGCGCGGTTGGACACTGTACGGCGACTGTGTGGGAGAAACAAAAGCCGGATGTTTCTGACAACGAAATATCATCGAAATTCGTGAAACTATTCCGAGGACTCAAAGATGTTAAAGACCTTATTAATGAGGAATATCTGGCTGCGAGCATCGAATCGGCGTGCCAGGATATCCAGTATCCTGCGATCGCGAAGATATGCAAAGACAACACGGCCCAATTCGAAAATTACATACATCACGTTCTCAAATCGAACACATCGGCCGAGACAATGTGCAAAATCGTCGGCATGTGTAACAACATGAAATTGGACAATATTATTTCGTTGAACAAGAAAAGCACCAACGTGCCCGTCAAGCATAAAGATCAGCTCCTTGGAAAATCCCGTTGTACTTGGGGTCCTTCGTATTGGTGCAGTAATTTTAGCACTGGCCGAGAATGCAACGCTACGCCTCACTGCATCAACCGCGTGTGGTCCAAAATGACCTTCCCGGAGGACAACGACAACGTATGCCAGATATGCTTGGACATGGTGAAGCAGGCGAGAGACCAACTACAGAGCAACGAAACTCAGGAAGAAATAAAAGAAGTATTCGAGGGTTCGTGCAAGTTGATTTCCATAAAGTTCGTGGCAGAGGGATGTATGAAATTGGCCGATGAGTTCGTGGTCGAGCTGATTGAGACGCTCGCTTCAGAGATGAACCCCCAAGCCGTGTGCTCTGTCGCTGGACTCTGTAACAACGCTAAGATCGATCGGCTGCTTGTAGACTACAACGCCCAAAGAGAACTAAGGGCCGGTTGTTACAATTGTCAGAAGACCGTCGGTGTTGTCAGGAAGAAGTTTGACGAGACCAAGTATGAAGACTTTTTAGTGGGACTCCTGCAGGTATGCCGCAACATGGACTCATTATCCGACTCTTGTTCGATGCTGATATTCAAATACTACGAGAACATTTTGGAAGCCGTGAAAAAGGATTTAAACCCCGAAGGCATCTGCCATGTCAGCGGACAGTGTTCGTACAAGTTCCATAATCACGATGAGTTTACGTTCCCTGAACAGATGGTCCAGTATTCTGCGACCGAAGATGTTCCGTGCGAATTTTGCGAACAGCTGGTCAAACATCTGCGAGACGTTCTCGTCGCTAATACTACTGAACTCGAATTCTATAAAGTCCTTCAAGGCCTCTGCAAGCAAACGGGAAAATTCAAGGACGAATGTCTACACTTAGCCGAACAGTACTACCCGGTTATCTACAACTTCCTAGTGTCCGATTTGAAACCGGCTGAAACGTGCAAAATGATTGGGATCTGCGGTAACCTCACTAGCGCGCCGATCTCGCCGCTCGTGGCCAGGGAACTGGTCGTTAAAGTGCAACCTAAATTGATTGGCGCTGAAGAATCAAAGATCGCTCATGTACCGTTAGCAAAACAAGCGGAGCCGGCGAGCGCGGCGGTGCCCGTGCTCCCCCTCGAGCGCATGTTCGTAGCGGCGCCGCAGAGCAAGGCCGCCTGCGCGTTCTGCCAGTACTTCCTGCACTACTTGCAGGTGCAACTCAGCGACACCAGGACCGAGGACAAAGTGAAGGCCGCAGTAGAGGAGGCGTGCGACGTGCTGCCCGACGCCCTCAACGGTGAATGCAAGGAGTTCGTAACGCAGTACGGATCCGCCGTCATCGCTTTGCTCGTTCAAGAAATCGATCCCGCCAGCGTATGTCCCGCTCTTCAAATCTGCCCTCAAACAGAAGAAATCCGCCGAGTCGACGTCAATTCTGAAAAATCTAACTGCCCCCTGTGCCTCTTCGCCGTTGAACAACTAGAATCGGTGCTCAAGAACAACCGTAGTGAGGAGAACATTCGCAAGGCTCTGGACGGTCTCTGCACGCGGCTCTCCCAGAAACTGCAGAGCGAGTGCATCGACTTCGTGGACACGTACTCGAGTCAGCTCGTCGAGATGCTGGTGGCCGACATGAACGCGAAGGAGATCTGCGTGTTCCTGAAGCTGTGTCGGGACCAACTGCACGATCCCCTCAAGCTGACCCACTCGTCCATAGACAAGTTCCACGCGAAGCCGACATTGAGAGGCGACAGGAACAACCACAGGAAGAAGTCGCTTCTGCCCAAACACATGCTGGTTTCTGAATTCAGTGACGTCGAAACGAACGAGATCCTGGACGACACGGTGAACGGTCGCCGCGTCTCGCACAAGTCTCAGAGCAACGTCTGTGTTCTTTGCGAATTCGTCCTGAAGGAAATCGACGACCAAATAAAAGACAAACATAACGACGACGAAATCAAGAAAGCTGTTCACGGCATATGCAAACACATGCCGAAGTCGGTCAGCGCTGAGTGCGACCAGTTCGTTGAGAAATACGCGGATTTAGTCATCTCTCTCCTCGCTCAAGAATTGGACCCGAGCGAAGTCTGCGAAGAATTGAAATTATGCAAACCAGAATCACTGAAAATTAATAAAATAAAAAAAGACATCCTCGACTGCGCCGTCTGCGAGACAGTGGTGATGGCCGTGAAGAAGGTCCTCAAAAACGAGAAGCTCGACCGCAACATCGTGCACATCATCGAGAAGTCGTGTGGACTTTTGCCCGCCAAATACAACGCTCAGTGTCACGCAATGCTGGAGATTTACGGCGAAAGCATCATTCACCTCATCGAAGAGTTTGGCGCGAAGGGCGTCTGCCAGAGGATCGGATTTTGTGGTCGGACCGAAACCGCATACGTGCGCATGTACCGCGAGAAGAGGAACTAA
Protein
MTNTFAVCLLSLTFLCCTNLSFARQVPKECAKGPQVWCESLKRGAECGAVGHCTATVWEKQKPDVSDNEISSKFVKLFRGLKDVKDLINEEYLAASIESACQDIQYPAIAKICKDNTAQFENYIHHVLKSNTSAETMCKIVGMCNNMKLDNIISLNKKSTNVPVKHKDQLLGKSRCTWGPSYWCSNFSTGRECNATPHCINRVWSKMTFPEDNDNVCQICLDMVKQARDQLQSNETQEEIKEVFEGSCKLISIKFVAEGCMKLADEFVVELIETLASEMNPQAVCSVAGLCNNAKIDRLLVDYNAQRELRAGCYNCQKTVGVVRKKFDETKYEDFLVGLLQVCRNMDSLSDSCSMLIFKYYENILEAVKKDLNPEGICHVSGQCSYKFHNHDEFTFPEQMVQYSATEDVPCEFCEQLVKHLRDVLVANTTELEFYKVLQGLCKQTGKFKDECLHLAEQYYPVIYNFLVSDLKPAETCKMIGICGNLTSAPISPLVARELVVKVQPKLIGAEESKIAHVPLAKQAEPASAAVPVLPLERMFVAAPQSKAACAFCQYFLHYLQVQLSDTRTEDKVKAAVEEACDVLPDALNGECKEFVTQYGSAVIALLVQEIDPASVCPALQICPQTEEIRRVDVNSEKSNCPLCLFAVEQLESVLKNNRSEENIRKALDGLCTRLSQKLQSECIDFVDTYSSQLVEMLVADMNAKEICVFLKLCRDQLHDPLKLTHSSIDKFHAKPTLRGDRNNHRKKSLLPKHMLVSEFSDVETNEILDDTVNGRRVSHKSQSNVCVLCEFVLKEIDDQIKDKHNDDEIKKAVHGICKHMPKSVSAECDQFVEKYADLVISLLAQELDPSEVCEELKLCKPESLKINKIKKDILDCAVCETVVMAVKKVLKNEKLDRNIVHIIEKSCGLLPAKYNAQCHAMLEIYGESIIHLIEEFGAKGVCQRIGFCGRTETAYVRMYREKRN
Summary
Uniprot
E7E2K2
O15997
A0A2A4K7G2
A0A1E1WKP2
A0A1E1WGK9
A0A194PHI9
+ More
S4P7C1 A0A212EYI5 A0A0U3TC83 D6WA77 V5GQ62 A0A2P8YEX6 A0A2J7RAC4 A0A158NLF1 F4WW78 A0A195B1M4 A0A182RVQ3 A0A154NY99 E2B1B7 A0A182QQD3 A0A195E6M8 A0A1W6EW39 A0A0C9R481 A0A0C9QS25 E2B7E4 Q0IGB5 A0A195CYA7 A0A1Q3FMP1 A0A1Q3FMD0 A0A182FMM1 A0A1Q3FMJ5 A0A1Q3FMM3 A0A2M3YXK5 Q7PMW6 A0A1Q3FML8 A0A1Q3FMN5 A0A2M3YXL6 F5HM00 A0A182MI21 A0A182YH17 A0A1S4F3J7 A0A2M3ZYN9 A0A2M3ZYI0 A0A182MZT0 A0A1Q3FMG7 A0A2M4BBG0 A0A2M4BBH3 A0A2M4BBH9 A0A182J5K7 A0A026X0V3 E9IQG2 A0A3L8DR53 A0A084VFR7 B3MT70 W5JL43 A0A182W5X7 A0A310SFK6 A0A336LVF0 B3P7V2 A0A1I8PUM6 U5ENL5 B4K915 A0A0M4EEE5 B4HZW6 A0A067RE75 B4R2A9 U4UQ14 A0A1J1HNS1 N6UD23 A0A1I8PUJ9 B4PMM8 A0A1W4U6A4 V9IEL2 Q9Y125 B0X4W7 A0A088AG18 A0A1L8EE11 Q3YMU5 B4NH07 A0A1L8EE43 Q29CC4 A0A2A3ERF5 A0A182KDI5 A0A182T2U5 A0A0A1WLJ9 A0A0L0CR79 A0A1L8DZ96 A0A1L8DZ81 A0A1L8DZR3 B4LW88 A0A1I8N5V3 A0A3B0JEU6 W8AIW3 A0A1L8DZ98
S4P7C1 A0A212EYI5 A0A0U3TC83 D6WA77 V5GQ62 A0A2P8YEX6 A0A2J7RAC4 A0A158NLF1 F4WW78 A0A195B1M4 A0A182RVQ3 A0A154NY99 E2B1B7 A0A182QQD3 A0A195E6M8 A0A1W6EW39 A0A0C9R481 A0A0C9QS25 E2B7E4 Q0IGB5 A0A195CYA7 A0A1Q3FMP1 A0A1Q3FMD0 A0A182FMM1 A0A1Q3FMJ5 A0A1Q3FMM3 A0A2M3YXK5 Q7PMW6 A0A1Q3FML8 A0A1Q3FMN5 A0A2M3YXL6 F5HM00 A0A182MI21 A0A182YH17 A0A1S4F3J7 A0A2M3ZYN9 A0A2M3ZYI0 A0A182MZT0 A0A1Q3FMG7 A0A2M4BBG0 A0A2M4BBH3 A0A2M4BBH9 A0A182J5K7 A0A026X0V3 E9IQG2 A0A3L8DR53 A0A084VFR7 B3MT70 W5JL43 A0A182W5X7 A0A310SFK6 A0A336LVF0 B3P7V2 A0A1I8PUM6 U5ENL5 B4K915 A0A0M4EEE5 B4HZW6 A0A067RE75 B4R2A9 U4UQ14 A0A1J1HNS1 N6UD23 A0A1I8PUJ9 B4PMM8 A0A1W4U6A4 V9IEL2 Q9Y125 B0X4W7 A0A088AG18 A0A1L8EE11 Q3YMU5 B4NH07 A0A1L8EE43 Q29CC4 A0A2A3ERF5 A0A182KDI5 A0A182T2U5 A0A0A1WLJ9 A0A0L0CR79 A0A1L8DZ96 A0A1L8DZ81 A0A1L8DZR3 B4LW88 A0A1I8N5V3 A0A3B0JEU6 W8AIW3 A0A1L8DZ98
Pubmed
9611271
26354079
23622113
22118469
26694822
18362917
+ More
19820115 29403074 21347285 21719571 20798317 17510324 12364791 14747013 17210077 25244985 24508170 21282665 30249741 24438588 17994087 20920257 23761445 24845553 23537049 17550304 10731132 12537568 12537572 12537573 12537574 15917496 16110336 17569856 17569867 26109357 26109356 15632085 25830018 26108605 25315136 24495485
19820115 29403074 21347285 21719571 20798317 17510324 12364791 14747013 17210077 25244985 24508170 21282665 30249741 24438588 17994087 20920257 23761445 24845553 23537049 17550304 10731132 12537568 12537572 12537573 12537574 15917496 16110336 17569856 17569867 26109357 26109356 15632085 25830018 26108605 25315136 24495485
EMBL
HQ606141
ADU03994.1
AB008449
BAA23126.1
NWSH01000098
PCG79590.1
+ More
GDQN01003464 JAT87590.1 GDQN01005053 JAT86001.1 KQ459603 KPI92881.1 GAIX01004704 JAA87856.1 AGBW02011531 OWR46517.1 KU218679 ALX00059.1 KQ971312 EEZ98045.1 GALX01004714 JAB63752.1 PYGN01000651 PSN42806.1 NEVH01006574 PNF37788.1 ADTU01019356 GL888404 EGI61549.1 KQ976662 KYM78393.1 KQ434773 KZC03998.1 GL444841 EFN60517.1 AXCN02001406 KQ979568 KYN20746.1 KY563538 ARK19947.1 GBYB01011109 JAG80876.1 GBYB01006549 JAG76316.1 GL446164 EFN88386.1 CH477260 EAT45724.1 KQ977115 KYN05638.1 GFDL01006253 JAV28792.1 GFDL01006338 JAV28707.1 GFDL01006322 JAV28723.1 GFDL01006250 JAV28795.1 GGFM01000251 MBW21002.1 AAAB01008967 EAA13115.5 GFDL01006224 JAV28821.1 GFDL01006228 JAV28817.1 GGFM01000250 MBW21001.1 EGK97311.1 AXCM01002370 GGFK01000294 MBW33615.1 GGFK01000292 MBW33613.1 GFDL01006274 JAV28771.1 GGFJ01001244 MBW50385.1 GGFJ01001245 MBW50386.1 GGFJ01001246 MBW50387.1 KK107063 EZA61024.1 GL764790 EFZ17192.1 QOIP01000005 RLU22887.1 ATLV01012459 KE524793 KFB36811.1 CH902623 EDV30460.2 ADMH02000853 ETN64846.1 KQ768883 OAD53035.1 UFQT01000137 SSX20679.1 CH954182 EDV53010.1 GANO01000550 JAB59321.1 CH933806 EDW16612.2 CP012526 ALC46354.1 CH480819 EDW53573.1 KK852692 KDR18348.1 CM000364 EDX15065.1 KB632314 ERL92231.1 CVRI01000010 CRK89044.1 APGK01033400 KB740874 ENN78536.1 CM000160 EDW99162.1 JR039944 AEY58916.1 AF145647 AE014297 DQ062780 AAD38622.1 AAF57097.1 AAY56653.1 DS232357 EDS40582.1 GFDG01001839 JAV16960.1 DQ062781 AAY56654.1 CH964272 EDW84504.1 GFDG01001838 JAV16961.1 CM000070 EAL26722.2 KZ288197 PBC33709.1 GBXI01014736 JAC99555.1 JRES01000032 KNC34741.1 GFDF01002303 JAV11781.1 GFDF01002304 JAV11780.1 GFDF01002302 JAV11782.1 CH940650 EDW67622.2 OUUW01000005 SPP80857.1 GAMC01018110 JAB88445.1 GFDF01002328 JAV11756.1
GDQN01003464 JAT87590.1 GDQN01005053 JAT86001.1 KQ459603 KPI92881.1 GAIX01004704 JAA87856.1 AGBW02011531 OWR46517.1 KU218679 ALX00059.1 KQ971312 EEZ98045.1 GALX01004714 JAB63752.1 PYGN01000651 PSN42806.1 NEVH01006574 PNF37788.1 ADTU01019356 GL888404 EGI61549.1 KQ976662 KYM78393.1 KQ434773 KZC03998.1 GL444841 EFN60517.1 AXCN02001406 KQ979568 KYN20746.1 KY563538 ARK19947.1 GBYB01011109 JAG80876.1 GBYB01006549 JAG76316.1 GL446164 EFN88386.1 CH477260 EAT45724.1 KQ977115 KYN05638.1 GFDL01006253 JAV28792.1 GFDL01006338 JAV28707.1 GFDL01006322 JAV28723.1 GFDL01006250 JAV28795.1 GGFM01000251 MBW21002.1 AAAB01008967 EAA13115.5 GFDL01006224 JAV28821.1 GFDL01006228 JAV28817.1 GGFM01000250 MBW21001.1 EGK97311.1 AXCM01002370 GGFK01000294 MBW33615.1 GGFK01000292 MBW33613.1 GFDL01006274 JAV28771.1 GGFJ01001244 MBW50385.1 GGFJ01001245 MBW50386.1 GGFJ01001246 MBW50387.1 KK107063 EZA61024.1 GL764790 EFZ17192.1 QOIP01000005 RLU22887.1 ATLV01012459 KE524793 KFB36811.1 CH902623 EDV30460.2 ADMH02000853 ETN64846.1 KQ768883 OAD53035.1 UFQT01000137 SSX20679.1 CH954182 EDV53010.1 GANO01000550 JAB59321.1 CH933806 EDW16612.2 CP012526 ALC46354.1 CH480819 EDW53573.1 KK852692 KDR18348.1 CM000364 EDX15065.1 KB632314 ERL92231.1 CVRI01000010 CRK89044.1 APGK01033400 KB740874 ENN78536.1 CM000160 EDW99162.1 JR039944 AEY58916.1 AF145647 AE014297 DQ062780 AAD38622.1 AAF57097.1 AAY56653.1 DS232357 EDS40582.1 GFDG01001839 JAV16960.1 DQ062781 AAY56654.1 CH964272 EDW84504.1 GFDG01001838 JAV16961.1 CM000070 EAL26722.2 KZ288197 PBC33709.1 GBXI01014736 JAC99555.1 JRES01000032 KNC34741.1 GFDF01002303 JAV11781.1 GFDF01002304 JAV11780.1 GFDF01002302 JAV11782.1 CH940650 EDW67622.2 OUUW01000005 SPP80857.1 GAMC01018110 JAB88445.1 GFDF01002328 JAV11756.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000007266
UP000245037
UP000235965
+ More
UP000005205 UP000007755 UP000078540 UP000075900 UP000076502 UP000000311 UP000075886 UP000078492 UP000008237 UP000008820 UP000078542 UP000069272 UP000007062 UP000075883 UP000076408 UP000075884 UP000075880 UP000053097 UP000279307 UP000030765 UP000007801 UP000000673 UP000075920 UP000008711 UP000095300 UP000009192 UP000092553 UP000001292 UP000027135 UP000000304 UP000030742 UP000183832 UP000019118 UP000002282 UP000192221 UP000000803 UP000002320 UP000005203 UP000007798 UP000001819 UP000242457 UP000075881 UP000075901 UP000037069 UP000008792 UP000095301 UP000268350
UP000005205 UP000007755 UP000078540 UP000075900 UP000076502 UP000000311 UP000075886 UP000078492 UP000008237 UP000008820 UP000078542 UP000069272 UP000007062 UP000075883 UP000076408 UP000075884 UP000075880 UP000053097 UP000279307 UP000030765 UP000007801 UP000000673 UP000075920 UP000008711 UP000095300 UP000009192 UP000092553 UP000001292 UP000027135 UP000000304 UP000030742 UP000183832 UP000019118 UP000002282 UP000192221 UP000000803 UP000002320 UP000005203 UP000007798 UP000001819 UP000242457 UP000075881 UP000075901 UP000037069 UP000008792 UP000095301 UP000268350
Interpro
ProteinModelPortal
E7E2K2
O15997
A0A2A4K7G2
A0A1E1WKP2
A0A1E1WGK9
A0A194PHI9
+ More
S4P7C1 A0A212EYI5 A0A0U3TC83 D6WA77 V5GQ62 A0A2P8YEX6 A0A2J7RAC4 A0A158NLF1 F4WW78 A0A195B1M4 A0A182RVQ3 A0A154NY99 E2B1B7 A0A182QQD3 A0A195E6M8 A0A1W6EW39 A0A0C9R481 A0A0C9QS25 E2B7E4 Q0IGB5 A0A195CYA7 A0A1Q3FMP1 A0A1Q3FMD0 A0A182FMM1 A0A1Q3FMJ5 A0A1Q3FMM3 A0A2M3YXK5 Q7PMW6 A0A1Q3FML8 A0A1Q3FMN5 A0A2M3YXL6 F5HM00 A0A182MI21 A0A182YH17 A0A1S4F3J7 A0A2M3ZYN9 A0A2M3ZYI0 A0A182MZT0 A0A1Q3FMG7 A0A2M4BBG0 A0A2M4BBH3 A0A2M4BBH9 A0A182J5K7 A0A026X0V3 E9IQG2 A0A3L8DR53 A0A084VFR7 B3MT70 W5JL43 A0A182W5X7 A0A310SFK6 A0A336LVF0 B3P7V2 A0A1I8PUM6 U5ENL5 B4K915 A0A0M4EEE5 B4HZW6 A0A067RE75 B4R2A9 U4UQ14 A0A1J1HNS1 N6UD23 A0A1I8PUJ9 B4PMM8 A0A1W4U6A4 V9IEL2 Q9Y125 B0X4W7 A0A088AG18 A0A1L8EE11 Q3YMU5 B4NH07 A0A1L8EE43 Q29CC4 A0A2A3ERF5 A0A182KDI5 A0A182T2U5 A0A0A1WLJ9 A0A0L0CR79 A0A1L8DZ96 A0A1L8DZ81 A0A1L8DZR3 B4LW88 A0A1I8N5V3 A0A3B0JEU6 W8AIW3 A0A1L8DZ98
S4P7C1 A0A212EYI5 A0A0U3TC83 D6WA77 V5GQ62 A0A2P8YEX6 A0A2J7RAC4 A0A158NLF1 F4WW78 A0A195B1M4 A0A182RVQ3 A0A154NY99 E2B1B7 A0A182QQD3 A0A195E6M8 A0A1W6EW39 A0A0C9R481 A0A0C9QS25 E2B7E4 Q0IGB5 A0A195CYA7 A0A1Q3FMP1 A0A1Q3FMD0 A0A182FMM1 A0A1Q3FMJ5 A0A1Q3FMM3 A0A2M3YXK5 Q7PMW6 A0A1Q3FML8 A0A1Q3FMN5 A0A2M3YXL6 F5HM00 A0A182MI21 A0A182YH17 A0A1S4F3J7 A0A2M3ZYN9 A0A2M3ZYI0 A0A182MZT0 A0A1Q3FMG7 A0A2M4BBG0 A0A2M4BBH3 A0A2M4BBH9 A0A182J5K7 A0A026X0V3 E9IQG2 A0A3L8DR53 A0A084VFR7 B3MT70 W5JL43 A0A182W5X7 A0A310SFK6 A0A336LVF0 B3P7V2 A0A1I8PUM6 U5ENL5 B4K915 A0A0M4EEE5 B4HZW6 A0A067RE75 B4R2A9 U4UQ14 A0A1J1HNS1 N6UD23 A0A1I8PUJ9 B4PMM8 A0A1W4U6A4 V9IEL2 Q9Y125 B0X4W7 A0A088AG18 A0A1L8EE11 Q3YMU5 B4NH07 A0A1L8EE43 Q29CC4 A0A2A3ERF5 A0A182KDI5 A0A182T2U5 A0A0A1WLJ9 A0A0L0CR79 A0A1L8DZ96 A0A1L8DZ81 A0A1L8DZR3 B4LW88 A0A1I8N5V3 A0A3B0JEU6 W8AIW3 A0A1L8DZ98
PDB
5U85
E-value=3.28341e-08,
Score=143
Ontologies
GO
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.998335
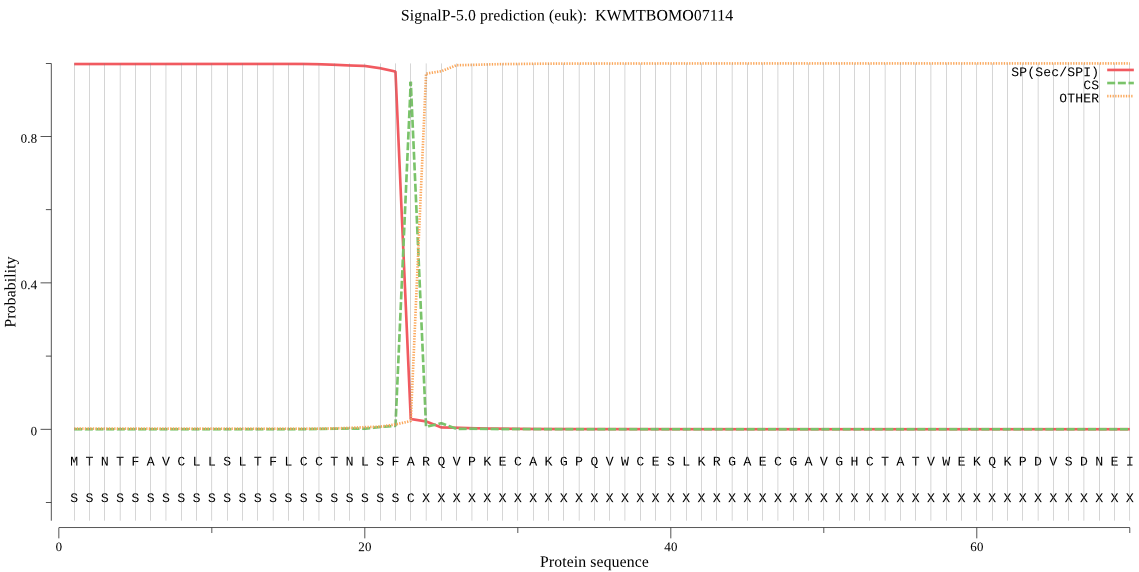
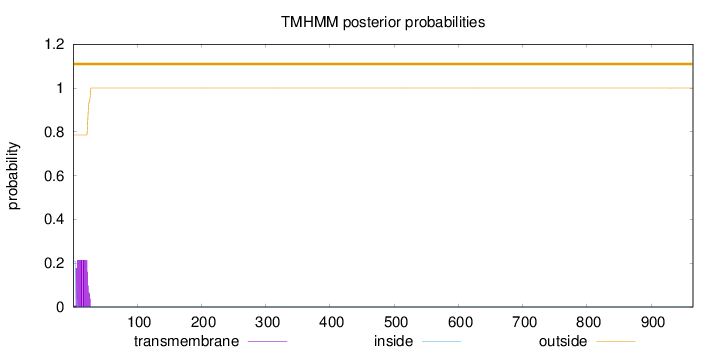
Length:
965
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.22454999999999
Exp number, first 60 AAs:
4.22247
Total prob of N-in:
0.21401
outside
1 - 965
Population Genetic Test Statistics
Pi
187.397363
Theta
153.748644
Tajima's D
-1.773926
CLR
0.421656
CSRT
0.0288985550722464
Interpretation
Possibly Positive selection
