Gene
KWMTBOMO07113
Pre Gene Modal
BGIBMGA010540
Annotation
PREDICTED:_nucleoside_diphosphate_kinase_homolog_5-like_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.624
Sequence
CDS
ATGTCCGTGACTTCATCGGACTCACAAGCACCTAGTCATGCGTATTCCGAACGCACGCTTGCATTGATTAAGCCGGAAGCATTCCAGGATTCAGAAGCTATTCAAGATCATATAAGAAACAATGGCTTCACTATCCTAGCTAAGCGCGTAGTTCAACTAACACCGGAGCAAGCAGCGGAGCTTTACAGAGGTCACTACGGACGACAACACTTTCCTCATTTAGTAGCTCAAATGTCATGCGGTCCGATAGTGGTATTGGTCTTAGCTGCCCGAGATTGCATTCAAAAATGGCGTACCCTTATGGGCCCGGCCAGGGTCGCCGAAGCTCAAGCATACTGGCCAGATAGTCTACGGGCATGCTACGGTCGTCGCACTAAATACGGAGATTATTTTAACGGTCTCCATGGCAGCGAGAACCTCGCGGAGGCTGTGAGAGAAATACATTTCTTCTTTCCTAACATGATAGTCGGTCCTTTACTTCGGAATTGGCAAATAAACGATTATATGCAGAAGTACATTACTCCCACGCTGCTGCCTGGCCTCACGGCTCTGGCTCACGAGCGACCCGCTGAACCAATATTATGGCTGGCAGAGTTTCTCCAACGTCACAATCCCAATGAACCGAAACTAGCCGATAAGATAAATCATCAGCGCGAACAACAGCGCTGCTACACTCCCGTGCCATCTGAAGAGAGTATTTTAAAGTAA
Protein
MSVTSSDSQAPSHAYSERTLALIKPEAFQDSEAIQDHIRNNGFTILAKRVVQLTPEQAAELYRGHYGRQHFPHLVAQMSCGPIVVLVLAARDCIQKWRTLMGPARVAEAQAYWPDSLRACYGRRTKYGDYFNGLHGSENLAEAVREIHFFFPNMIVGPLLRNWQINDYMQKYITPTLLPGLTALAHERPAEPILWLAEFLQRHNPNEPKLADKINHQREQQRCYTPVPSEESILK
Summary
Similarity
Belongs to the NDK family.
Uniprot
H9JLY8
A0A2A4K698
A0A212EYE5
A0A194PHT9
A0A194QTT1
A0A2W1BLJ1
+ More
A0A2H1W375 A0A0L7LGS6 R7VDT5 A0A026WC64 A0A0M9A460 A0A3S0ZQV8 A0A3L8DQ32 A0A2J7RGB8 A0A2P8ZDQ8 A0A0B7BLA3 H2RMP2 A0A088AQ63 A0A195B5N2 A0A1I8J6H0 A0A1I8J7U1 C3Z4Q2 V4AX29 A0A154P751 A0A3P8VUU1 A0A2C9JFT3 A0A1S3HVK6 H2RMP3 A0A3B4DIH6 A0A1I8FZ90 A0A3B4BQQ4 A0A3M6TTD0 E3TFQ7 A0A3Q1FKB2 Q5C0E8 C1LEL0 A0A0J7L453 A0A210QX70 A0A151I935 A0A3Q2ZSG0 A0A310SAD2 A0A151WKK2 A0A195F349 A0A091HQS0 A0A2B4SG78 A0A2Y9IN21 A0A1U7TI02 A0A3B4TJR5 K1R1K7 Q6DGQ8 E3TC92 A0A3Q0SPW4 A7SAT1 E4WZV0 A0A195EDH3 A0A3B4X0M7 A0A2Y9I609 A0A3B1JW45 F1R4T6 A0A3Q0SPY4 A0A1V4KUS0 A0A3P9C455 A0A3P8P3Q6 A0A2I0MA10 D2HK89 A0A3P8P3X7 A0A3P8P3E9 E2RLI0 A0A3Q7SB01 A0A384C4I7 A0A3Q7XBR5 A0A3P8P3E2 T2MEZ8 G1U1P7 A0A3Q3EJD0 G1PFK6 A0A0P7VNB2 A0A060Y8Z4 A0A3Q3LEJ0 A0A3Q3EJB1 M3YE61 A0A096MRH7 U3J2N0 A0A3Q7NXI8 A0A2U3Y0L8 A0A1S2ZDA8 A0A3Q1CHN4 A0A2U3VRP5 A0A099ZYX1 A0A3B4EPN7 G7P8D5 G7MUV9 A0A1I8H3N6 A0A2K6BWU4 A0A3B3BIB8 H9GNT0 A0A2K5RV07 B5XCD3 A0A1S3SUU9 A0A3B4ERJ6
A0A2H1W375 A0A0L7LGS6 R7VDT5 A0A026WC64 A0A0M9A460 A0A3S0ZQV8 A0A3L8DQ32 A0A2J7RGB8 A0A2P8ZDQ8 A0A0B7BLA3 H2RMP2 A0A088AQ63 A0A195B5N2 A0A1I8J6H0 A0A1I8J7U1 C3Z4Q2 V4AX29 A0A154P751 A0A3P8VUU1 A0A2C9JFT3 A0A1S3HVK6 H2RMP3 A0A3B4DIH6 A0A1I8FZ90 A0A3B4BQQ4 A0A3M6TTD0 E3TFQ7 A0A3Q1FKB2 Q5C0E8 C1LEL0 A0A0J7L453 A0A210QX70 A0A151I935 A0A3Q2ZSG0 A0A310SAD2 A0A151WKK2 A0A195F349 A0A091HQS0 A0A2B4SG78 A0A2Y9IN21 A0A1U7TI02 A0A3B4TJR5 K1R1K7 Q6DGQ8 E3TC92 A0A3Q0SPW4 A7SAT1 E4WZV0 A0A195EDH3 A0A3B4X0M7 A0A2Y9I609 A0A3B1JW45 F1R4T6 A0A3Q0SPY4 A0A1V4KUS0 A0A3P9C455 A0A3P8P3Q6 A0A2I0MA10 D2HK89 A0A3P8P3X7 A0A3P8P3E9 E2RLI0 A0A3Q7SB01 A0A384C4I7 A0A3Q7XBR5 A0A3P8P3E2 T2MEZ8 G1U1P7 A0A3Q3EJD0 G1PFK6 A0A0P7VNB2 A0A060Y8Z4 A0A3Q3LEJ0 A0A3Q3EJB1 M3YE61 A0A096MRH7 U3J2N0 A0A3Q7NXI8 A0A2U3Y0L8 A0A1S2ZDA8 A0A3Q1CHN4 A0A2U3VRP5 A0A099ZYX1 A0A3B4EPN7 G7P8D5 G7MUV9 A0A1I8H3N6 A0A2K6BWU4 A0A3B3BIB8 H9GNT0 A0A2K5RV07 B5XCD3 A0A1S3SUU9 A0A3B4ERJ6
Pubmed
19121390
22118469
26354079
28756777
26227816
23254933
+ More
24508170 30249741 29403074 21551351 26392545 18563158 24487278 15562597 30382153 20634964 16617374 28812685 22992520 17615350 21097902 25329095 23594743 25186727 23371554 20010809 16341006 24813606 24065732 21993624 24755649 23749191 22002653 17431167 29451363 21881562 20433749
24508170 30249741 29403074 21551351 26392545 18563158 24487278 15562597 30382153 20634964 16617374 28812685 22992520 17615350 21097902 25329095 23594743 25186727 23371554 20010809 16341006 24813606 24065732 21993624 24755649 23749191 22002653 17431167 29451363 21881562 20433749
EMBL
BABH01005455
NWSH01000098
PCG79589.1
AGBW02011531
OWR46516.1
KQ459603
+ More
KPI92882.1 KQ461194 KPJ06951.1 KZ149975 PZC75952.1 ODYU01005902 SOQ47272.1 JTDY01001252 KOB74401.1 AMQN01004214 KB292914 ELU16784.1 KK107293 EZA53261.1 KQ435739 KOX76865.1 RQTK01000233 RUS83718.1 QOIP01000005 RLU22396.1 NEVH01004401 PNF39884.1 PYGN01000087 PSN54634.1 HACG01046863 CEK93728.1 KQ976598 KYM79580.1 NIVC01001097 NIVC01001059 PAA72339.1 PAA72806.1 NIVC01000225 PAA87565.1 GG666580 EEN52491.1 KB201004 ESO99615.1 KQ434829 KZC07765.1 RCHS01002970 RMX44655.1 GU589191 ADO29143.1 AY810988 AAX26877.2 FN317407 CAX73138.1 LBMM01000892 KMQ97264.1 NEDP02001389 OWF53324.1 KQ978323 KYM95140.1 KQ766694 OAD53591.1 KQ983002 KYQ48409.1 KQ981855 KYN34888.1 KL217647 KFO97442.1 LSMT01000090 PFX28053.1 JH817943 EKC43147.1 BC076282 AAH76282.1 GU587971 ADO27928.1 DS469611 EDO39201.1 FN653019 FN657060 CBY22696.1 CBY42344.1 KQ979039 KYN23258.1 FQ377590 LSYS01001584 OPJ88152.1 AKCR02000025 PKK26515.1 ACTA01003696 ACTA01179687 ACTA01187685 ACTA01195683 GL192940 EFB18537.1 AAEX03007811 AAEX03007812 HAAD01004437 CDG70669.1 AAGW02027777 AAGW02027778 AAGW02027779 AAPE02012945 JARO02001602 KPP74946.1 FR908440 CDQ88191.1 AEYP01016737 AEYP01016738 AEYP01016739 AEYP01016740 AEYP01016741 AHZZ02030049 AHZZ02030050 ADON01130636 KL870149 KGL87011.1 AQIA01060619 AQIA01060620 AQIA01060621 AQIA01060622 AQIA01060623 AQIA01060624 AQIA01060625 CM001281 EHH54556.1 JSUE03037003 JSUE03037004 JSUE03037005 CM001258 EHH26824.1 NIVC01000300 PAA85954.1 AAWZ02036183 BT048702 ACI68503.1
KPI92882.1 KQ461194 KPJ06951.1 KZ149975 PZC75952.1 ODYU01005902 SOQ47272.1 JTDY01001252 KOB74401.1 AMQN01004214 KB292914 ELU16784.1 KK107293 EZA53261.1 KQ435739 KOX76865.1 RQTK01000233 RUS83718.1 QOIP01000005 RLU22396.1 NEVH01004401 PNF39884.1 PYGN01000087 PSN54634.1 HACG01046863 CEK93728.1 KQ976598 KYM79580.1 NIVC01001097 NIVC01001059 PAA72339.1 PAA72806.1 NIVC01000225 PAA87565.1 GG666580 EEN52491.1 KB201004 ESO99615.1 KQ434829 KZC07765.1 RCHS01002970 RMX44655.1 GU589191 ADO29143.1 AY810988 AAX26877.2 FN317407 CAX73138.1 LBMM01000892 KMQ97264.1 NEDP02001389 OWF53324.1 KQ978323 KYM95140.1 KQ766694 OAD53591.1 KQ983002 KYQ48409.1 KQ981855 KYN34888.1 KL217647 KFO97442.1 LSMT01000090 PFX28053.1 JH817943 EKC43147.1 BC076282 AAH76282.1 GU587971 ADO27928.1 DS469611 EDO39201.1 FN653019 FN657060 CBY22696.1 CBY42344.1 KQ979039 KYN23258.1 FQ377590 LSYS01001584 OPJ88152.1 AKCR02000025 PKK26515.1 ACTA01003696 ACTA01179687 ACTA01187685 ACTA01195683 GL192940 EFB18537.1 AAEX03007811 AAEX03007812 HAAD01004437 CDG70669.1 AAGW02027777 AAGW02027778 AAGW02027779 AAPE02012945 JARO02001602 KPP74946.1 FR908440 CDQ88191.1 AEYP01016737 AEYP01016738 AEYP01016739 AEYP01016740 AEYP01016741 AHZZ02030049 AHZZ02030050 ADON01130636 KL870149 KGL87011.1 AQIA01060619 AQIA01060620 AQIA01060621 AQIA01060622 AQIA01060623 AQIA01060624 AQIA01060625 CM001281 EHH54556.1 JSUE03037003 JSUE03037004 JSUE03037005 CM001258 EHH26824.1 NIVC01000300 PAA85954.1 AAWZ02036183 BT048702 ACI68503.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000014760 UP000053097 UP000053105 UP000271974 UP000279307 UP000235965 UP000245037 UP000005226 UP000005203 UP000078540 UP000095280 UP000215902 UP000001554 UP000030746 UP000076502 UP000265120 UP000076420 UP000085678 UP000261440 UP000275408 UP000221080 UP000257200 UP000036403 UP000242188 UP000078542 UP000264800 UP000075809 UP000078541 UP000054308 UP000225706 UP000248482 UP000189704 UP000261420 UP000005408 UP000261340 UP000001593 UP000078492 UP000261360 UP000248481 UP000018467 UP000000437 UP000190648 UP000265160 UP000265100 UP000053872 UP000008912 UP000002254 UP000286640 UP000261680 UP000291021 UP000286642 UP000001811 UP000261660 UP000001074 UP000034805 UP000192224 UP000193380 UP000000715 UP000028761 UP000016666 UP000286641 UP000245341 UP000079721 UP000257160 UP000245340 UP000053858 UP000261460 UP000009130 UP000233100 UP000006718 UP000233120 UP000261560 UP000001646 UP000233040 UP000087266
UP000014760 UP000053097 UP000053105 UP000271974 UP000279307 UP000235965 UP000245037 UP000005226 UP000005203 UP000078540 UP000095280 UP000215902 UP000001554 UP000030746 UP000076502 UP000265120 UP000076420 UP000085678 UP000261440 UP000275408 UP000221080 UP000257200 UP000036403 UP000242188 UP000078542 UP000264800 UP000075809 UP000078541 UP000054308 UP000225706 UP000248482 UP000189704 UP000261420 UP000005408 UP000261340 UP000001593 UP000078492 UP000261360 UP000248481 UP000018467 UP000000437 UP000190648 UP000265160 UP000265100 UP000053872 UP000008912 UP000002254 UP000286640 UP000261680 UP000291021 UP000286642 UP000001811 UP000261660 UP000001074 UP000034805 UP000192224 UP000193380 UP000000715 UP000028761 UP000016666 UP000286641 UP000245341 UP000079721 UP000257160 UP000245340 UP000053858 UP000261460 UP000009130 UP000233100 UP000006718 UP000233120 UP000261560 UP000001646 UP000233040 UP000087266
PRIDE
Interpro
SUPFAM
SSF54919
SSF54919
Gene 3D
ProteinModelPortal
H9JLY8
A0A2A4K698
A0A212EYE5
A0A194PHT9
A0A194QTT1
A0A2W1BLJ1
+ More
A0A2H1W375 A0A0L7LGS6 R7VDT5 A0A026WC64 A0A0M9A460 A0A3S0ZQV8 A0A3L8DQ32 A0A2J7RGB8 A0A2P8ZDQ8 A0A0B7BLA3 H2RMP2 A0A088AQ63 A0A195B5N2 A0A1I8J6H0 A0A1I8J7U1 C3Z4Q2 V4AX29 A0A154P751 A0A3P8VUU1 A0A2C9JFT3 A0A1S3HVK6 H2RMP3 A0A3B4DIH6 A0A1I8FZ90 A0A3B4BQQ4 A0A3M6TTD0 E3TFQ7 A0A3Q1FKB2 Q5C0E8 C1LEL0 A0A0J7L453 A0A210QX70 A0A151I935 A0A3Q2ZSG0 A0A310SAD2 A0A151WKK2 A0A195F349 A0A091HQS0 A0A2B4SG78 A0A2Y9IN21 A0A1U7TI02 A0A3B4TJR5 K1R1K7 Q6DGQ8 E3TC92 A0A3Q0SPW4 A7SAT1 E4WZV0 A0A195EDH3 A0A3B4X0M7 A0A2Y9I609 A0A3B1JW45 F1R4T6 A0A3Q0SPY4 A0A1V4KUS0 A0A3P9C455 A0A3P8P3Q6 A0A2I0MA10 D2HK89 A0A3P8P3X7 A0A3P8P3E9 E2RLI0 A0A3Q7SB01 A0A384C4I7 A0A3Q7XBR5 A0A3P8P3E2 T2MEZ8 G1U1P7 A0A3Q3EJD0 G1PFK6 A0A0P7VNB2 A0A060Y8Z4 A0A3Q3LEJ0 A0A3Q3EJB1 M3YE61 A0A096MRH7 U3J2N0 A0A3Q7NXI8 A0A2U3Y0L8 A0A1S2ZDA8 A0A3Q1CHN4 A0A2U3VRP5 A0A099ZYX1 A0A3B4EPN7 G7P8D5 G7MUV9 A0A1I8H3N6 A0A2K6BWU4 A0A3B3BIB8 H9GNT0 A0A2K5RV07 B5XCD3 A0A1S3SUU9 A0A3B4ERJ6
A0A2H1W375 A0A0L7LGS6 R7VDT5 A0A026WC64 A0A0M9A460 A0A3S0ZQV8 A0A3L8DQ32 A0A2J7RGB8 A0A2P8ZDQ8 A0A0B7BLA3 H2RMP2 A0A088AQ63 A0A195B5N2 A0A1I8J6H0 A0A1I8J7U1 C3Z4Q2 V4AX29 A0A154P751 A0A3P8VUU1 A0A2C9JFT3 A0A1S3HVK6 H2RMP3 A0A3B4DIH6 A0A1I8FZ90 A0A3B4BQQ4 A0A3M6TTD0 E3TFQ7 A0A3Q1FKB2 Q5C0E8 C1LEL0 A0A0J7L453 A0A210QX70 A0A151I935 A0A3Q2ZSG0 A0A310SAD2 A0A151WKK2 A0A195F349 A0A091HQS0 A0A2B4SG78 A0A2Y9IN21 A0A1U7TI02 A0A3B4TJR5 K1R1K7 Q6DGQ8 E3TC92 A0A3Q0SPW4 A7SAT1 E4WZV0 A0A195EDH3 A0A3B4X0M7 A0A2Y9I609 A0A3B1JW45 F1R4T6 A0A3Q0SPY4 A0A1V4KUS0 A0A3P9C455 A0A3P8P3Q6 A0A2I0MA10 D2HK89 A0A3P8P3X7 A0A3P8P3E9 E2RLI0 A0A3Q7SB01 A0A384C4I7 A0A3Q7XBR5 A0A3P8P3E2 T2MEZ8 G1U1P7 A0A3Q3EJD0 G1PFK6 A0A0P7VNB2 A0A060Y8Z4 A0A3Q3LEJ0 A0A3Q3EJB1 M3YE61 A0A096MRH7 U3J2N0 A0A3Q7NXI8 A0A2U3Y0L8 A0A1S2ZDA8 A0A3Q1CHN4 A0A2U3VRP5 A0A099ZYX1 A0A3B4EPN7 G7P8D5 G7MUV9 A0A1I8H3N6 A0A2K6BWU4 A0A3B3BIB8 H9GNT0 A0A2K5RV07 B5XCD3 A0A1S3SUU9 A0A3B4ERJ6
PDB
3ZTS
E-value=2.01075e-17,
Score=215
Ontologies
GO
Topology
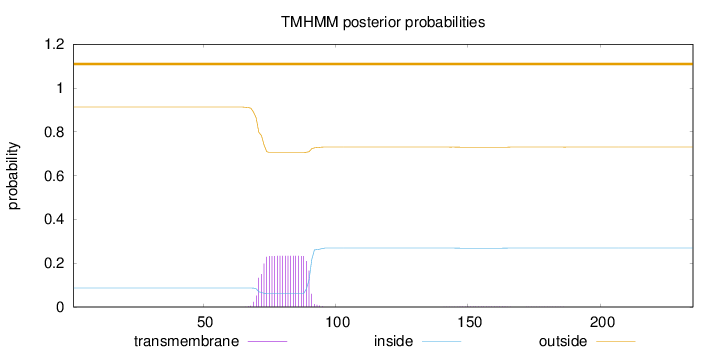
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.56945
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.08698
outside
1 - 235
Population Genetic Test Statistics
Pi
37.237607
Theta
189.514214
Tajima's D
1.123528
CLR
0.16516
CSRT
0.689965501724914
Interpretation
Uncertain
