Gene
KWMTBOMO07103 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010423
Annotation
cytosolic_malate_dehydrogenase_[Bombyx_mori]
Full name
Malate dehydrogenase
Location in the cell
Cytoplasmic Reliability : 2.022
Sequence
CDS
ATGGCTGAACCTATCAGAGTTGTTGTAACTGGTGCTGCCGGACAAATTGCATACTCACTTCTTTATCAAATTGCGTCTGGAGCAGTTTTTGGACCTCAGCAACCTGTCTTCCTCCACCTTCTTGATATTGCGCCTATGATGGGTGTACTTGAAGGTGTTGTCATGGAGTTGGCTGACTGTGCTCTGCCACTTTTGGCTGGGGTTCTTCCTACAGCAAATCCTGAAGAAGCTTTCAAAGATGTTGCTGCTGCTTTCCTAGTTGGTGCTATGCCCAGAAAGGAAGGTATGGAGAGGAAGGATCTTCTTGCTGCTAATGTGCGCATCTTCAAAGAGCAGGGCCAGGCTTTGGACAAAGTGGCTCGTAAAGATGTGAAGGTCCTTGTTGTTGGCAACCCAGCCAATACAAATGCTCTAATATGTTCTAAGTATGCTCCATCTATTCCAAAAGAAAATTTCACTGCCATGACTCGTCTCGATCAAAACAGGGCTCAGTCCCAACTTGCTGCTAAAATTGGAGTACCGGTTAAAGATGTTAAGAGAGTAATCATCTGGGGAAACCATTCATCAACCCAGTTCCCTGATGCTTCGAATGCTGTAGCTATTGTTGGAGGTGCTCAAAAATCTGTTTCTGAAATTATCAATGACGATGCATATTTGAAAGGCGCATTTGTAACAACGGTTCAAAAACGTGGTGCTGCCGTTATTGCTGCCAGGAAAATGTCATCTGCTCTTTCTGCTGCTAAAGCTGCATCAGACCACATGAGAGATTGGTTCCTTGGCACTGAAGACAGATGGGTTAGCATGGGAGTTGTTTCAGATGGATCTTATGGTACTCCGCGTGATGTTGTATATTCATTCCCTGTTACTGTTACCAATGGAAAGTGGAAAATTGTTGAGGGTCTCACTATCTCTGATTTTGCCCGTCAAATGCTCGATGCTACTGGCAAAGAGTTGGTTGAAGAAAAACAGGAGGCTTTAGATGTCTGCAAAGATTGA
Protein
MAEPIRVVVTGAAGQIAYSLLYQIASGAVFGPQQPVFLHLLDIAPMMGVLEGVVMELADCALPLLAGVLPTANPEEAFKDVAAAFLVGAMPRKEGMERKDLLAANVRIFKEQGQALDKVARKDVKVLVVGNPANTNALICSKYAPSIPKENFTAMTRLDQNRAQSQLAAKIGVPVKDVKRVIIWGNHSSTQFPDASNAVAIVGGAQKSVSEIINDDAYLKGAFVTTVQKRGAAVIAARKMSSALSAAKAASDHMRDWFLGTEDRWVSMGVVSDGSYGTPRDVVYSFPVTVTNGKWKIVEGLTISDFARQMLDATGKELVEEKQEALDVCKD
Summary
Catalytic Activity
(S)-malate + NAD(+) = H(+) + NADH + oxaloacetate
Similarity
Belongs to the LDH/MDH superfamily.
Uniprot
Q2F5P8
H9JLM2
A0A2A4K6Q8
A0A2A4K5R7
A0A2W1BLI1
A0A2H1WV70
+ More
I3RRT0 A0A194PHJ8 I4DKD3 A0A194QPN6 A0A212EYD5 A0A220K8N0 A0A3S2P348 S4PWZ8 A0A0L7KT71 J3JTT8 D8VJF0 D8VJH0 D8VJD0 D8VJE2 D8VJI8 D8VJF1 D8VJH1 D8VJD9 A0A1W5YLL5 V5GL68 A0A023EPX1 A0A336LJF6 A0A336LI46 A0A336MYN4 A0A182GEL7 A0A182GAF8 A0A1S4FHF8 A0A0L0CHM9 B3MJG6 A0A0P5EX35 T1P7W6 A0A1L8EFR3 A0A1Q3FJK9 A0A1I8NGI3 A0A1I8NZF7 A0A0P5EZ88 A0A0P5P0A4 A0A1W4USV7 Q8MQS7 B3N9N7 Q9VKX2 B4NZN3 A0A0P5XNA2 A0A1A9V2A7 G3K809 G3K7Y9 B4HWN2 G3K811 A0A0P5Y320 A0A0P5L6B1 Q171B2 B4Q988 G3K7Z3 D3TLF0 A0A0P4XFV9 G3K804 G3K808 G3K7Z5 G3K801 A0A0P5GU82 G3K7Z9 G3K7Z6 A0A0P5GUI1 A0A0P5NVC8 G3K7Y7 A0A0P5JLW8 G3K7Z8 A0A1A9W5C1 G3K802 G3K7Z0 G3K807 G3K7Z2 G3K806 A0A0P5EUD8 A0A0N8A3T0 G3K800 G3K7Y8 A0A1B0A4T6 A0A034W6Y2 G3K7Z7 W8CCW0 E9HA96 R4WN61 B3TGZ5 G3K803 A0A0P4WVN4 A0A1D2NAR9 A0A0K8UJL7 A0A3B0JHE0 B4NQQ2 A0A0P5JLT4 A0A182IZK3 K7J699 A0A0A1WL85
I3RRT0 A0A194PHJ8 I4DKD3 A0A194QPN6 A0A212EYD5 A0A220K8N0 A0A3S2P348 S4PWZ8 A0A0L7KT71 J3JTT8 D8VJF0 D8VJH0 D8VJD0 D8VJE2 D8VJI8 D8VJF1 D8VJH1 D8VJD9 A0A1W5YLL5 V5GL68 A0A023EPX1 A0A336LJF6 A0A336LI46 A0A336MYN4 A0A182GEL7 A0A182GAF8 A0A1S4FHF8 A0A0L0CHM9 B3MJG6 A0A0P5EX35 T1P7W6 A0A1L8EFR3 A0A1Q3FJK9 A0A1I8NGI3 A0A1I8NZF7 A0A0P5EZ88 A0A0P5P0A4 A0A1W4USV7 Q8MQS7 B3N9N7 Q9VKX2 B4NZN3 A0A0P5XNA2 A0A1A9V2A7 G3K809 G3K7Y9 B4HWN2 G3K811 A0A0P5Y320 A0A0P5L6B1 Q171B2 B4Q988 G3K7Z3 D3TLF0 A0A0P4XFV9 G3K804 G3K808 G3K7Z5 G3K801 A0A0P5GU82 G3K7Z9 G3K7Z6 A0A0P5GUI1 A0A0P5NVC8 G3K7Y7 A0A0P5JLW8 G3K7Z8 A0A1A9W5C1 G3K802 G3K7Z0 G3K807 G3K7Z2 G3K806 A0A0P5EUD8 A0A0N8A3T0 G3K800 G3K7Y8 A0A1B0A4T6 A0A034W6Y2 G3K7Z7 W8CCW0 E9HA96 R4WN61 B3TGZ5 G3K803 A0A0P4WVN4 A0A1D2NAR9 A0A0K8UJL7 A0A3B0JHE0 B4NQQ2 A0A0P5JLT4 A0A182IZK3 K7J699 A0A0A1WL85
EC Number
1.1.1.37
Pubmed
19121390
28756777
26354079
22651552
22118469
28630352
+ More
23622113 26227816 22516182 23537049 20442862 24945155 26483478 26108605 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21841803 17510324 22936249 20353571 25348373 24495485 21292972 23691247 27289101 20075255 25830018
23622113 26227816 22516182 23537049 20442862 24945155 26483478 26108605 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21841803 17510324 22936249 20353571 25348373 24495485 21292972 23691247 27289101 20075255 25830018
EMBL
DQ311375
ABD36319.1
BABH01005443
NWSH01000098
PCG79594.1
PCG79595.1
+ More
KZ149975 PZC75942.1 ODYU01011308 SOQ56969.1 JQ730683 AFK28500.1 KQ459603 KPI92891.1 AK401751 BAM18373.1 KQ461194 KPJ06940.1 AGBW02011531 OWR46508.1 MF319729 ASJ26443.1 RSAL01002350 RVE40506.1 GAIX01005913 JAA86647.1 JTDY01006181 KOB66214.1 BT126647 KB632390 AEE61610.1 ERL94450.1 GQ987207 GQ987209 GQ987210 GQ987211 GQ987212 GQ987215 GQ987217 GQ987221 GQ987223 ADI81894.1 GQ987227 GQ987229 GQ987230 GQ987231 GQ987232 GQ987233 GQ987234 GQ987237 GQ987238 GQ987239 GQ987240 GQ987241 GQ987242 GQ987243 GQ987244 GQ987246 ADI81914.1 GQ987187 GQ987188 GQ987189 GQ987190 GQ987191 GQ987192 GQ987193 GQ987194 GQ987195 GQ987197 GQ987198 GQ987202 GQ987203 GQ987204 GQ987205 GQ987206 ADI81874.1 GQ987199 GQ987200 GQ987201 ADI81886.1 GQ987245 ADI81932.1 GQ987208 GQ987213 GQ987214 GQ987216 GQ987218 GQ987219 GQ987220 GQ987222 GQ987224 GQ987225 GQ987226 ADI81895.1 GQ987228 GQ987235 GQ987236 ADI81915.1 GQ987196 ADI81883.1 KX147676 ARI45080.1 GALX01003627 JAB64839.1 GAPW01002495 JAC11103.1 UFQS01005467 UFQT01005467 SSX16823.1 SSX36004.1 UFQS01004279 UFQT01004279 SSX16303.1 SSX35623.1 UFQT01003982 SSX35406.1 JXUM01058239 JXUM01058240 JXUM01058241 KQ561993 KXJ76965.1 JXUM01050947 JXUM01050948 JXUM01050949 KQ561669 KXJ77853.1 JRES01000384 KNC31756.1 CH902620 EDV32334.1 GDIQ01268852 JAJ82872.1 KA644721 AFP59350.1 GFDG01001237 JAV17562.1 GFDL01007379 JAV27666.1 GDIP01200441 GDIP01195646 GDIQ01270092 GDIQ01199368 GDIQ01182332 GDIP01088100 GDIP01037918 GDIQ01049131 LRGB01003024 JAJ22961.1 JAJ81632.1 KZS05014.1 GDIQ01142046 JAL09680.1 AY128413 AAM75006.1 CH954177 EDV58532.1 AE014134 BT128780 AAF52935.2 AEK87152.1 CM000157 EDW88817.1 GDIP01071211 JAM32504.1 JN174131 JN174132 AEN67674.1 AEN67675.1 JN174111 JN174113 AEN67654.1 AEN67656.1 CH480818 EDW52427.1 JN174133 AEN67676.1 GDIP01063712 JAM40003.1 GDIQ01199367 JAK52358.1 CH477460 EAT40580.1 CM000361 CM002910 EDX04530.1 KMY89518.1 JN174115 AEN67658.1 EZ422252 ADD18528.1 GDIP01251443 JAI71958.1 JN174126 AEN67669.1 JN174130 AEN67673.1 JN174116 JN174117 AEN67659.1 AEN67660.1 JN174123 AEN67666.1 GDIQ01235557 JAK16168.1 JN174121 AEN67664.1 JN174118 AEN67661.1 GDIQ01242410 JAK09315.1 GDIQ01158727 JAK92998.1 JN174109 AEN67652.1 GDIQ01196572 JAK55153.1 JN174120 AEN67663.1 JN174124 AEN67667.1 JN174112 AEN67655.1 JN174129 AEN67672.1 JN174114 AEN67657.1 JN174128 AEN67671.1 GDIQ01270040 JAJ81684.1 GDIP01185297 JAJ38105.1 JN174122 AEN67665.1 JN174110 AEN67653.1 GAKP01009066 GAKP01009057 JAC49895.1 JN174119 AEN67662.1 GAMC01003011 JAC03545.1 GL732611 EFX71334.1 AK417036 BAN20251.1 EU141617 ABX57441.1 JN174125 AEN67668.1 GDIP01251444 JAI71957.1 LJIJ01000115 ODN02348.1 GDHF01025412 JAI26902.1 OUUW01000006 SPP81605.1 CH964295 EDW86691.2 GDIQ01203069 JAK48656.1 GBXI01015124 JAC99167.1
KZ149975 PZC75942.1 ODYU01011308 SOQ56969.1 JQ730683 AFK28500.1 KQ459603 KPI92891.1 AK401751 BAM18373.1 KQ461194 KPJ06940.1 AGBW02011531 OWR46508.1 MF319729 ASJ26443.1 RSAL01002350 RVE40506.1 GAIX01005913 JAA86647.1 JTDY01006181 KOB66214.1 BT126647 KB632390 AEE61610.1 ERL94450.1 GQ987207 GQ987209 GQ987210 GQ987211 GQ987212 GQ987215 GQ987217 GQ987221 GQ987223 ADI81894.1 GQ987227 GQ987229 GQ987230 GQ987231 GQ987232 GQ987233 GQ987234 GQ987237 GQ987238 GQ987239 GQ987240 GQ987241 GQ987242 GQ987243 GQ987244 GQ987246 ADI81914.1 GQ987187 GQ987188 GQ987189 GQ987190 GQ987191 GQ987192 GQ987193 GQ987194 GQ987195 GQ987197 GQ987198 GQ987202 GQ987203 GQ987204 GQ987205 GQ987206 ADI81874.1 GQ987199 GQ987200 GQ987201 ADI81886.1 GQ987245 ADI81932.1 GQ987208 GQ987213 GQ987214 GQ987216 GQ987218 GQ987219 GQ987220 GQ987222 GQ987224 GQ987225 GQ987226 ADI81895.1 GQ987228 GQ987235 GQ987236 ADI81915.1 GQ987196 ADI81883.1 KX147676 ARI45080.1 GALX01003627 JAB64839.1 GAPW01002495 JAC11103.1 UFQS01005467 UFQT01005467 SSX16823.1 SSX36004.1 UFQS01004279 UFQT01004279 SSX16303.1 SSX35623.1 UFQT01003982 SSX35406.1 JXUM01058239 JXUM01058240 JXUM01058241 KQ561993 KXJ76965.1 JXUM01050947 JXUM01050948 JXUM01050949 KQ561669 KXJ77853.1 JRES01000384 KNC31756.1 CH902620 EDV32334.1 GDIQ01268852 JAJ82872.1 KA644721 AFP59350.1 GFDG01001237 JAV17562.1 GFDL01007379 JAV27666.1 GDIP01200441 GDIP01195646 GDIQ01270092 GDIQ01199368 GDIQ01182332 GDIP01088100 GDIP01037918 GDIQ01049131 LRGB01003024 JAJ22961.1 JAJ81632.1 KZS05014.1 GDIQ01142046 JAL09680.1 AY128413 AAM75006.1 CH954177 EDV58532.1 AE014134 BT128780 AAF52935.2 AEK87152.1 CM000157 EDW88817.1 GDIP01071211 JAM32504.1 JN174131 JN174132 AEN67674.1 AEN67675.1 JN174111 JN174113 AEN67654.1 AEN67656.1 CH480818 EDW52427.1 JN174133 AEN67676.1 GDIP01063712 JAM40003.1 GDIQ01199367 JAK52358.1 CH477460 EAT40580.1 CM000361 CM002910 EDX04530.1 KMY89518.1 JN174115 AEN67658.1 EZ422252 ADD18528.1 GDIP01251443 JAI71958.1 JN174126 AEN67669.1 JN174130 AEN67673.1 JN174116 JN174117 AEN67659.1 AEN67660.1 JN174123 AEN67666.1 GDIQ01235557 JAK16168.1 JN174121 AEN67664.1 JN174118 AEN67661.1 GDIQ01242410 JAK09315.1 GDIQ01158727 JAK92998.1 JN174109 AEN67652.1 GDIQ01196572 JAK55153.1 JN174120 AEN67663.1 JN174124 AEN67667.1 JN174112 AEN67655.1 JN174129 AEN67672.1 JN174114 AEN67657.1 JN174128 AEN67671.1 GDIQ01270040 JAJ81684.1 GDIP01185297 JAJ38105.1 JN174122 AEN67665.1 JN174110 AEN67653.1 GAKP01009066 GAKP01009057 JAC49895.1 JN174119 AEN67662.1 GAMC01003011 JAC03545.1 GL732611 EFX71334.1 AK417036 BAN20251.1 EU141617 ABX57441.1 JN174125 AEN67668.1 GDIP01251444 JAI71957.1 LJIJ01000115 ODN02348.1 GDHF01025412 JAI26902.1 OUUW01000006 SPP81605.1 CH964295 EDW86691.2 GDIQ01203069 JAK48656.1 GBXI01015124 JAC99167.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000030742 UP000069940 UP000249989 UP000037069 UP000007801 UP000095301 UP000095300 UP000076858 UP000192221 UP000008711 UP000000803 UP000002282 UP000078200 UP000001292 UP000008820 UP000000304 UP000091820 UP000092445 UP000000305 UP000094527 UP000268350 UP000007798 UP000075880 UP000002358
UP000037510 UP000030742 UP000069940 UP000249989 UP000037069 UP000007801 UP000095301 UP000095300 UP000076858 UP000192221 UP000008711 UP000000803 UP000002282 UP000078200 UP000001292 UP000008820 UP000000304 UP000091820 UP000092445 UP000000305 UP000094527 UP000268350 UP000007798 UP000075880 UP000002358
Interpro
IPR015955
Lactate_DH/Glyco_Ohase_4_C
+ More
IPR001557 L-lactate/malate_DH
IPR010945 Malate_DH_type2
IPR022383 Lactate/malate_DH_C
IPR011274 Malate_DH_NAD-dep_euk
IPR036291 NAD(P)-bd_dom_sf
IPR001252 Malate_DH_AS
IPR001236 Lactate/malate_DH_N
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR001557 L-lactate/malate_DH
IPR010945 Malate_DH_type2
IPR022383 Lactate/malate_DH_C
IPR011274 Malate_DH_NAD-dep_euk
IPR036291 NAD(P)-bd_dom_sf
IPR001252 Malate_DH_AS
IPR001236 Lactate/malate_DH_N
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
Gene 3D
ProteinModelPortal
Q2F5P8
H9JLM2
A0A2A4K6Q8
A0A2A4K5R7
A0A2W1BLI1
A0A2H1WV70
+ More
I3RRT0 A0A194PHJ8 I4DKD3 A0A194QPN6 A0A212EYD5 A0A220K8N0 A0A3S2P348 S4PWZ8 A0A0L7KT71 J3JTT8 D8VJF0 D8VJH0 D8VJD0 D8VJE2 D8VJI8 D8VJF1 D8VJH1 D8VJD9 A0A1W5YLL5 V5GL68 A0A023EPX1 A0A336LJF6 A0A336LI46 A0A336MYN4 A0A182GEL7 A0A182GAF8 A0A1S4FHF8 A0A0L0CHM9 B3MJG6 A0A0P5EX35 T1P7W6 A0A1L8EFR3 A0A1Q3FJK9 A0A1I8NGI3 A0A1I8NZF7 A0A0P5EZ88 A0A0P5P0A4 A0A1W4USV7 Q8MQS7 B3N9N7 Q9VKX2 B4NZN3 A0A0P5XNA2 A0A1A9V2A7 G3K809 G3K7Y9 B4HWN2 G3K811 A0A0P5Y320 A0A0P5L6B1 Q171B2 B4Q988 G3K7Z3 D3TLF0 A0A0P4XFV9 G3K804 G3K808 G3K7Z5 G3K801 A0A0P5GU82 G3K7Z9 G3K7Z6 A0A0P5GUI1 A0A0P5NVC8 G3K7Y7 A0A0P5JLW8 G3K7Z8 A0A1A9W5C1 G3K802 G3K7Z0 G3K807 G3K7Z2 G3K806 A0A0P5EUD8 A0A0N8A3T0 G3K800 G3K7Y8 A0A1B0A4T6 A0A034W6Y2 G3K7Z7 W8CCW0 E9HA96 R4WN61 B3TGZ5 G3K803 A0A0P4WVN4 A0A1D2NAR9 A0A0K8UJL7 A0A3B0JHE0 B4NQQ2 A0A0P5JLT4 A0A182IZK3 K7J699 A0A0A1WL85
I3RRT0 A0A194PHJ8 I4DKD3 A0A194QPN6 A0A212EYD5 A0A220K8N0 A0A3S2P348 S4PWZ8 A0A0L7KT71 J3JTT8 D8VJF0 D8VJH0 D8VJD0 D8VJE2 D8VJI8 D8VJF1 D8VJH1 D8VJD9 A0A1W5YLL5 V5GL68 A0A023EPX1 A0A336LJF6 A0A336LI46 A0A336MYN4 A0A182GEL7 A0A182GAF8 A0A1S4FHF8 A0A0L0CHM9 B3MJG6 A0A0P5EX35 T1P7W6 A0A1L8EFR3 A0A1Q3FJK9 A0A1I8NGI3 A0A1I8NZF7 A0A0P5EZ88 A0A0P5P0A4 A0A1W4USV7 Q8MQS7 B3N9N7 Q9VKX2 B4NZN3 A0A0P5XNA2 A0A1A9V2A7 G3K809 G3K7Y9 B4HWN2 G3K811 A0A0P5Y320 A0A0P5L6B1 Q171B2 B4Q988 G3K7Z3 D3TLF0 A0A0P4XFV9 G3K804 G3K808 G3K7Z5 G3K801 A0A0P5GU82 G3K7Z9 G3K7Z6 A0A0P5GUI1 A0A0P5NVC8 G3K7Y7 A0A0P5JLW8 G3K7Z8 A0A1A9W5C1 G3K802 G3K7Z0 G3K807 G3K7Z2 G3K806 A0A0P5EUD8 A0A0N8A3T0 G3K800 G3K7Y8 A0A1B0A4T6 A0A034W6Y2 G3K7Z7 W8CCW0 E9HA96 R4WN61 B3TGZ5 G3K803 A0A0P4WVN4 A0A1D2NAR9 A0A0K8UJL7 A0A3B0JHE0 B4NQQ2 A0A0P5JLT4 A0A182IZK3 K7J699 A0A0A1WL85
PDB
5MDH
E-value=6.2306e-111,
Score=1024
Ontologies
KEGG
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
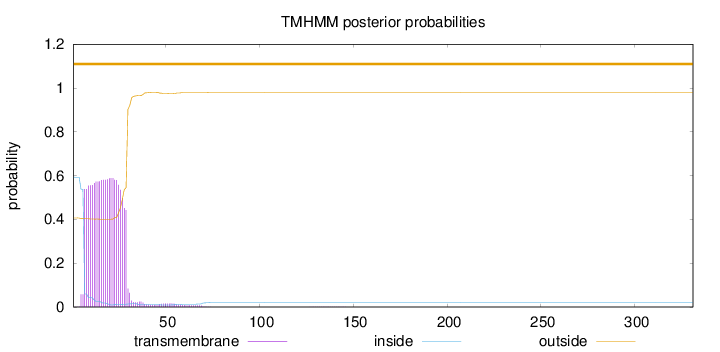
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.54362
Exp number, first 60 AAs:
13.46683
Total prob of N-in:
0.59303
POSSIBLE N-term signal
sequence
outside
1 - 331
Population Genetic Test Statistics
Pi
218.888333
Theta
217.417974
Tajima's D
-0.2695
CLR
46.919453
CSRT
0.296435178241088
Interpretation
Uncertain
