Gene
KWMTBOMO07101 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010536
Annotation
PREDICTED:_lactase-phlorizin_hydrolase-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.091 PlasmaMembrane Reliability : 1.549
Sequence
CDS
ATGAAGTTTGAACATCGGATTATCTCGGAAGAAGCAGTGACAATGCTTCATTTACCTTCCAGTTATTTATTAGCATTATTAATATTGTTATGTATTTTAACGAATGCAACTTCGCGCGAATTCCCTAAAGGATTTTTGTTTGGAGCTGCTACAGCCTCTTACCAAATCGAAGGCGCTTGGAATATTGACGGAAAAACTGAAAATATATGGGATCGTGCCTGTCACAAAGTACCCAGCCCGGTAGCCGATAACTCTACGGGAGACATCGCAAATGATTCCTATAAACTTTACAAACGAGATGTTGAAATGATAAGAGAACTTGGTGTTGATTATTACAGATTCTCTGTGTCCTGGACTCGTATTTTACCCACGAGTTTCCCAGATTACATAAATGAAGCTGGAGTTGCTTATTACAATAATCTGATTGACGAATTACTCAAATACAATATTGAGCCGATGCTCACAATTTACCACTGGGACTTACCACAGAAATTACAGGATATGGGAGGTTGGACGAACCCATATATTATCGATTGGTATGCGGACTACGCCAGAGTTCTGTTCCAAAAATTTGGTGATCGAGTTAAGAATTGGATCACTATAAACGAACCGAAAGAAATTTGCTTCCAAGGTTACGCAATAGGTACATTAGCTCCAGCATACACTATGAGCGGAGTGGCTGATTACTTGTGTGGGAAAAACGTTCTTTTAGCTCACGCTAAAGCTTATCATATCTATGACAAAGAATTCCGACCTACCCAAAATGGAAATATCTCTATAACCATTAGTTGCCAATGGTATGAGCCTGAAAGCGATCGCGAAGAAGATATTAACGCAGCGAATGAAATGATCTTATTCGAGTGTGGTCAATACGGTCACCCTATATTTACTGACAGCGGTGACTATCCCGAAATAGTCAAGCAACGTGTAGCAGCTAAGAGTGCTGAACAAGGTTTCTTACGCTCACGGTTCCCAGAGCTAAGCCCTGAAGAAGTTCAATACATCCGTGGAACATCAGACTTTTTTGGTTTAAATCACTACTCGACATTTTTGACGTACAGAAATGAGTCAGTTGTCGGCTACTACGAAATACCTTCGTATAATGATGATGTTGGAGTACTCTTTTATGACAAGGAGGAATGGTCCATTGGTTCGCATAACAGAGTTAAGACCACACCATGGGGCTTCTACAAACTTTTGAGACAAATTTCGAATTTTTACAATAACGTTCCGATTATAATCACTGAAAACGGTTTCGCGACCCTTACCGGCTTCGAAGACAACGACCGAATCACGCATCTGAGATTGTATTTAAATGCTCTTCTAGACGCTATTAAAGATGGATCCGATATCAGACTGTATACTCCTTGGAGCTTGATGGATAATTTTGAATGGATGTCTGGATATACGCAAAGATTTGGACTTTATGAAGTTGATTACGACAGTCCAGAACTTACACGGACGCCGAGAAAATCTGCTTTTATATTCAAAGAAATTGTAAAATCAAGGACTCTAGACTTTGATTATAAACCTGAAAGCTACGTTATGACGATTGACGAAGGAAAATGA
Protein
MKFEHRIISEEAVTMLHLPSSYLLALLILLCILTNATSREFPKGFLFGAATASYQIEGAWNIDGKTENIWDRACHKVPSPVADNSTGDIANDSYKLYKRDVEMIRELGVDYYRFSVSWTRILPTSFPDYINEAGVAYYNNLIDELLKYNIEPMLTIYHWDLPQKLQDMGGWTNPYIIDWYADYARVLFQKFGDRVKNWITINEPKEICFQGYAIGTLAPAYTMSGVADYLCGKNVLLAHAKAYHIYDKEFRPTQNGNISITISCQWYEPESDREEDINAANEMILFECGQYGHPIFTDSGDYPEIVKQRVAAKSAEQGFLRSRFPELSPEEVQYIRGTSDFFGLNHYSTFLTYRNESVVGYYEIPSYNDDVGVLFYDKEEWSIGSHNRVKTTPWGFYKLLRQISNFYNNVPIIITENGFATLTGFEDNDRITHLRLYLNALLDAIKDGSDIRLYTPWSLMDNFEWMSGYTQRFGLYEVDYDSPELTRTPRKSAFIFKEIVKSRTLDFDYKPESYVMTIDEGK
Summary
Similarity
Belongs to the glycosyl hydrolase 1 family.
Uniprot
H9JLY4
H9JLY5
A0A2A4JPS5
A0A194R3H3
A0A194R360
A0A194PMT2
+ More
A0A194PPG9 A0A194PPG4 A0A2H1VF65 A0A194R3H6 A0A194R4S7 A0A194R365 A0A194PNR8 A0A194PMT8 A0A194R8P7 A0A194PNS3 G9F9G9 G9F9I3 A0A3S2PE71 A0A194R400 A0A212EXF5 A0A212FF90 A0A212FF93 A0A194PU18 A0A194R3Z4 A0A0L7KVS3 A0A0L7LEZ3 A0A194PMG8 A0A212FBG8 A0A212EXF2 H9JXB3 A0A3S2NZ22 G9F9H3 A0A2W1BGQ3 A0A2H1VVL0 A0A2W1BA28 A0A2A4JQ50 A0A2A4JS29 A0A212FEI2 A0A2H1VWZ6 A0A2H1WY61 A0A2A4JNV2 A0A2A4JNT8 O61594 H9IZH5 A0A194RQF8 H9JMQ9 A0A2A4JPU6 A0A194PJU3 A0A2A4JCQ8 A0A2H1VWN4 A0A212F963 A0A1Y9IW90 A0A2H1VWC6 A0A194PRE4 H9J7W0 A0A194QNZ7 A0A2A4JPN8 A0A194PVV9 A0A182LWK9 A0A2W1BQ65 A0A0N1IQ38 A0A2W1BPZ1 A0A1Y9GKS9 A0NES8 A0A1Y9IRG1 A0A1Y9J0G9 A0A194R738 A0A2A4JTU1 A0A182LL19 A0A182J2M6 A0A2W1BWF3 A0A212F3C5 B0WNN7 A0A3S2M0F9 A0A2H1VWQ1 A0A1Y9GKT0 Q5TQY8 V5GZG1 A0A1Y9IRY8 Q16WF2 A0A182U0B4 A0A182QNT7 A0A194QDR0 A0A2H1W4S5 A0A1S4G3W5 A0A182HAK9 A0A182YP84 A0A182G2R0 A0A182HAK7 A0A182LL18 Q16ET6 A0A212ERQ2 Q7PI98 A0A182TWE4 A0A1Y9IS75 A0A1L8DQN1 Q16WF4 A0A1W1EGM5 A0A182YP82
A0A194PPG9 A0A194PPG4 A0A2H1VF65 A0A194R3H6 A0A194R4S7 A0A194R365 A0A194PNR8 A0A194PMT8 A0A194R8P7 A0A194PNS3 G9F9G9 G9F9I3 A0A3S2PE71 A0A194R400 A0A212EXF5 A0A212FF90 A0A212FF93 A0A194PU18 A0A194R3Z4 A0A0L7KVS3 A0A0L7LEZ3 A0A194PMG8 A0A212FBG8 A0A212EXF2 H9JXB3 A0A3S2NZ22 G9F9H3 A0A2W1BGQ3 A0A2H1VVL0 A0A2W1BA28 A0A2A4JQ50 A0A2A4JS29 A0A212FEI2 A0A2H1VWZ6 A0A2H1WY61 A0A2A4JNV2 A0A2A4JNT8 O61594 H9IZH5 A0A194RQF8 H9JMQ9 A0A2A4JPU6 A0A194PJU3 A0A2A4JCQ8 A0A2H1VWN4 A0A212F963 A0A1Y9IW90 A0A2H1VWC6 A0A194PRE4 H9J7W0 A0A194QNZ7 A0A2A4JPN8 A0A194PVV9 A0A182LWK9 A0A2W1BQ65 A0A0N1IQ38 A0A2W1BPZ1 A0A1Y9GKS9 A0NES8 A0A1Y9IRG1 A0A1Y9J0G9 A0A194R738 A0A2A4JTU1 A0A182LL19 A0A182J2M6 A0A2W1BWF3 A0A212F3C5 B0WNN7 A0A3S2M0F9 A0A2H1VWQ1 A0A1Y9GKT0 Q5TQY8 V5GZG1 A0A1Y9IRY8 Q16WF2 A0A182U0B4 A0A182QNT7 A0A194QDR0 A0A2H1W4S5 A0A1S4G3W5 A0A182HAK9 A0A182YP84 A0A182G2R0 A0A182HAK7 A0A182LL18 Q16ET6 A0A212ERQ2 Q7PI98 A0A182TWE4 A0A1Y9IS75 A0A1L8DQN1 Q16WF4 A0A1W1EGM5 A0A182YP82
Pubmed
EMBL
BABH01005437
BABH01005438
NWSH01000819
PCG74065.1
KQ460779
KPJ12242.1
+ More
KPJ12243.1 KQ459598 KPI94627.1 KPI94629.1 KPI94624.1 ODYU01001971 SOQ38904.1 KPJ12247.1 KPJ12245.1 KPJ12248.1 KPI94628.1 KPI94632.1 KPJ12246.1 KPI94633.1 JN033714 AEW46867.1 JN033728 AEW46881.1 RSAL01000077 RVE48784.1 KPJ12249.1 AGBW02011758 OWR46180.1 AGBW02008847 OWR52398.1 OWR52397.1 KPI94630.1 KPJ12244.1 JTDY01005139 KOB67338.1 JTDY01001400 KOB73970.1 KPI94631.1 AGBW02009344 OWR51063.1 OWR46178.1 BABH01035726 RSAL01000019 RVE52748.1 JN033718 AEW46871.1 KZ150146 PZC72895.1 ODYU01004710 SOQ44881.1 KZ150738 PZC70347.1 NWSH01000883 PCG73724.1 NWSH01000679 PCG74847.1 AGBW02008931 OWR52151.1 ODYU01004903 SOQ45256.1 ODYU01011923 SOQ57942.1 PCG73727.1 PCG73725.1 AF052729 AAC06038.1 BABH01014157 KQ459875 KPJ19634.1 BABH01016738 PCG73728.1 KQ459601 KPI93701.1 NWSH01001860 PCG69887.1 SOQ45257.1 AGBW02009638 OWR50268.1 ODYU01004836 SOQ45125.1 KPI93700.1 BABH01020023 BABH01020024 KQ458761 KPJ05206.1 PCG73726.1 KQ459589 KPI97521.1 AXCM01009805 KZ149936 PZC77128.1 KQ459993 KPJ18573.1 PZC77129.1 APCN01002262 AAAB01008960 EAU76599.1 KQ460870 KPJ11681.1 NWSH01000598 PCG75431.1 PZC77130.1 AGBW02010585 OWR48224.1 DS232013 EDS31873.1 RSAL01000086 RVE48281.1 SOQ45255.1 EAL40075.2 GALX01002583 JAB65883.1 CH477568 EAT38910.1 AXCN02000455 KQ459144 KPJ03597.1 ODYU01006202 SOQ47842.1 JXUM01031354 KQ560919 KXJ80350.1 JXUM01040976 KQ561266 KXJ79155.1 JXUM01031352 KXJ80348.1 CH478582 EAT32750.1 AGBW02012985 OWR44134.1 EAA44227.3 GFDF01005306 JAV08778.1 EAT38908.1 LT635664 SGZ49394.1
KPJ12243.1 KQ459598 KPI94627.1 KPI94629.1 KPI94624.1 ODYU01001971 SOQ38904.1 KPJ12247.1 KPJ12245.1 KPJ12248.1 KPI94628.1 KPI94632.1 KPJ12246.1 KPI94633.1 JN033714 AEW46867.1 JN033728 AEW46881.1 RSAL01000077 RVE48784.1 KPJ12249.1 AGBW02011758 OWR46180.1 AGBW02008847 OWR52398.1 OWR52397.1 KPI94630.1 KPJ12244.1 JTDY01005139 KOB67338.1 JTDY01001400 KOB73970.1 KPI94631.1 AGBW02009344 OWR51063.1 OWR46178.1 BABH01035726 RSAL01000019 RVE52748.1 JN033718 AEW46871.1 KZ150146 PZC72895.1 ODYU01004710 SOQ44881.1 KZ150738 PZC70347.1 NWSH01000883 PCG73724.1 NWSH01000679 PCG74847.1 AGBW02008931 OWR52151.1 ODYU01004903 SOQ45256.1 ODYU01011923 SOQ57942.1 PCG73727.1 PCG73725.1 AF052729 AAC06038.1 BABH01014157 KQ459875 KPJ19634.1 BABH01016738 PCG73728.1 KQ459601 KPI93701.1 NWSH01001860 PCG69887.1 SOQ45257.1 AGBW02009638 OWR50268.1 ODYU01004836 SOQ45125.1 KPI93700.1 BABH01020023 BABH01020024 KQ458761 KPJ05206.1 PCG73726.1 KQ459589 KPI97521.1 AXCM01009805 KZ149936 PZC77128.1 KQ459993 KPJ18573.1 PZC77129.1 APCN01002262 AAAB01008960 EAU76599.1 KQ460870 KPJ11681.1 NWSH01000598 PCG75431.1 PZC77130.1 AGBW02010585 OWR48224.1 DS232013 EDS31873.1 RSAL01000086 RVE48281.1 SOQ45255.1 EAL40075.2 GALX01002583 JAB65883.1 CH477568 EAT38910.1 AXCN02000455 KQ459144 KPJ03597.1 ODYU01006202 SOQ47842.1 JXUM01031354 KQ560919 KXJ80350.1 JXUM01040976 KQ561266 KXJ79155.1 JXUM01031352 KXJ80348.1 CH478582 EAT32750.1 AGBW02012985 OWR44134.1 EAA44227.3 GFDF01005306 JAV08778.1 EAT38908.1 LT635664 SGZ49394.1
Proteomes
Interpro
SUPFAM
SSF51445
SSF51445
ProteinModelPortal
H9JLY4
H9JLY5
A0A2A4JPS5
A0A194R3H3
A0A194R360
A0A194PMT2
+ More
A0A194PPG9 A0A194PPG4 A0A2H1VF65 A0A194R3H6 A0A194R4S7 A0A194R365 A0A194PNR8 A0A194PMT8 A0A194R8P7 A0A194PNS3 G9F9G9 G9F9I3 A0A3S2PE71 A0A194R400 A0A212EXF5 A0A212FF90 A0A212FF93 A0A194PU18 A0A194R3Z4 A0A0L7KVS3 A0A0L7LEZ3 A0A194PMG8 A0A212FBG8 A0A212EXF2 H9JXB3 A0A3S2NZ22 G9F9H3 A0A2W1BGQ3 A0A2H1VVL0 A0A2W1BA28 A0A2A4JQ50 A0A2A4JS29 A0A212FEI2 A0A2H1VWZ6 A0A2H1WY61 A0A2A4JNV2 A0A2A4JNT8 O61594 H9IZH5 A0A194RQF8 H9JMQ9 A0A2A4JPU6 A0A194PJU3 A0A2A4JCQ8 A0A2H1VWN4 A0A212F963 A0A1Y9IW90 A0A2H1VWC6 A0A194PRE4 H9J7W0 A0A194QNZ7 A0A2A4JPN8 A0A194PVV9 A0A182LWK9 A0A2W1BQ65 A0A0N1IQ38 A0A2W1BPZ1 A0A1Y9GKS9 A0NES8 A0A1Y9IRG1 A0A1Y9J0G9 A0A194R738 A0A2A4JTU1 A0A182LL19 A0A182J2M6 A0A2W1BWF3 A0A212F3C5 B0WNN7 A0A3S2M0F9 A0A2H1VWQ1 A0A1Y9GKT0 Q5TQY8 V5GZG1 A0A1Y9IRY8 Q16WF2 A0A182U0B4 A0A182QNT7 A0A194QDR0 A0A2H1W4S5 A0A1S4G3W5 A0A182HAK9 A0A182YP84 A0A182G2R0 A0A182HAK7 A0A182LL18 Q16ET6 A0A212ERQ2 Q7PI98 A0A182TWE4 A0A1Y9IS75 A0A1L8DQN1 Q16WF4 A0A1W1EGM5 A0A182YP82
A0A194PPG9 A0A194PPG4 A0A2H1VF65 A0A194R3H6 A0A194R4S7 A0A194R365 A0A194PNR8 A0A194PMT8 A0A194R8P7 A0A194PNS3 G9F9G9 G9F9I3 A0A3S2PE71 A0A194R400 A0A212EXF5 A0A212FF90 A0A212FF93 A0A194PU18 A0A194R3Z4 A0A0L7KVS3 A0A0L7LEZ3 A0A194PMG8 A0A212FBG8 A0A212EXF2 H9JXB3 A0A3S2NZ22 G9F9H3 A0A2W1BGQ3 A0A2H1VVL0 A0A2W1BA28 A0A2A4JQ50 A0A2A4JS29 A0A212FEI2 A0A2H1VWZ6 A0A2H1WY61 A0A2A4JNV2 A0A2A4JNT8 O61594 H9IZH5 A0A194RQF8 H9JMQ9 A0A2A4JPU6 A0A194PJU3 A0A2A4JCQ8 A0A2H1VWN4 A0A212F963 A0A1Y9IW90 A0A2H1VWC6 A0A194PRE4 H9J7W0 A0A194QNZ7 A0A2A4JPN8 A0A194PVV9 A0A182LWK9 A0A2W1BQ65 A0A0N1IQ38 A0A2W1BPZ1 A0A1Y9GKS9 A0NES8 A0A1Y9IRG1 A0A1Y9J0G9 A0A194R738 A0A2A4JTU1 A0A182LL19 A0A182J2M6 A0A2W1BWF3 A0A212F3C5 B0WNN7 A0A3S2M0F9 A0A2H1VWQ1 A0A1Y9GKT0 Q5TQY8 V5GZG1 A0A1Y9IRY8 Q16WF2 A0A182U0B4 A0A182QNT7 A0A194QDR0 A0A2H1W4S5 A0A1S4G3W5 A0A182HAK9 A0A182YP84 A0A182G2R0 A0A182HAK7 A0A182LL18 Q16ET6 A0A212ERQ2 Q7PI98 A0A182TWE4 A0A1Y9IS75 A0A1L8DQN1 Q16WF4 A0A1W1EGM5 A0A182YP82
PDB
5CG0
E-value=2.83103e-150,
Score=1365
Ontologies
GO
PANTHER
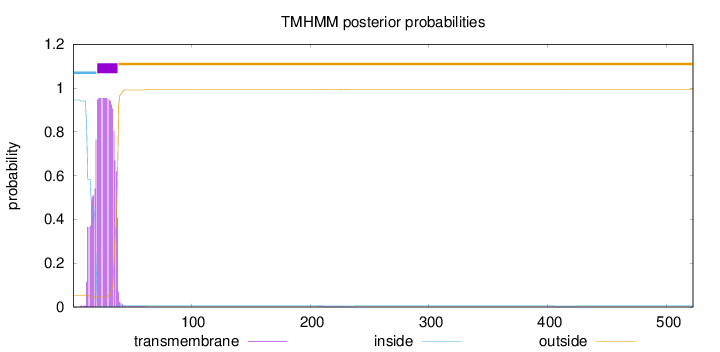
Topology
Length:
522
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.0273799999999
Exp number, first 60 AAs:
19.95739
Total prob of N-in:
0.94590
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 38
outside
39 - 522
Population Genetic Test Statistics
Pi
161.833177
Theta
128.732694
Tajima's D
0.514757
CLR
0.015139
CSRT
0.51852407379631
Interpretation
Uncertain
