Gene
KWMTBOMO07097
Annotation
PREDICTED:_lachesin-like_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.776 PlasmaMembrane Reliability : 1.478
Sequence
CDS
ATGGCGCGCACTGCGGGTGCCGTCTGCTTGTTCACGGTTAATTTGTTTTCCTGCATTATTGTTTGTCGTCAAGACAGTGGTAATAATCGGCCTCGAGATCAGAGCACGGTTTCTGAATCGTCAAGGAGATTTCGGACCGAGTTCTTAGAGGATTGGCCTCAAGCCCCTGGCGCACCATACTTCGATCCCAGTACTCCGGACAACGTCACGGGACTTGTGGGACATCCTGTGACTCTTCTGTGTAAAGTAAAGAATCTTCAGAATAGAACGGTATCATGGGTGAGGCATCGAGATATCCATTTGCTGACTGTGGGACGATATACTTACACGTCGGATCAGCGCTTCGAGGCGCAGCACAAGCCGCGCTCCGAGGAGTGGGCACTTCGAATCCGCAGTCCACAGCGACGCGACTCCGGCCAGTACGAGTGTCAGATCTCGACCACACCACCCATCGGACATGCTGTCTTCCTCAATATTGTGGAACCCGAGACAGAAGTATTAGGTGGACCAGATATATTTATTTACGCTGGTTCCACTATAAACCTCACTTGCATTGTTCGGCACACTCCAGAACCTCCAAGTACAATTAACTGGTCACATCGAGGGAAGACGATAAACTTCGACTCGACCCGCGGTGGGATATCGTTGGTGACCGAGAAAGGTGTTCATAGTAGCAGCCGACTATTAGTACAAGCTGCCCGCACGACCGACGCAGGCGCGTATCAATGCGCACCAGACAACGCACAGTCAGCTACTGCAAGAGTTCATGTGCTTACGGGAGAAAGTCCAGCGGCGATGCAGGGTAGTGCTCACTGGGCGGACAGATTCCGGCTACCAACACTCATCTTCTTTGCAACCATCGTATACCTAACAATATTGATACCTTGA
Protein
MARTAGAVCLFTVNLFSCIIVCRQDSGNNRPRDQSTVSESSRRFRTEFLEDWPQAPGAPYFDPSTPDNVTGLVGHPVTLLCKVKNLQNRTVSWVRHRDIHLLTVGRYTYTSDQRFEAQHKPRSEEWALRIRSPQRRDSGQYECQISTTPPIGHAVFLNIVEPETEVLGGPDIFIYAGSTINLTCIVRHTPEPPSTINWSHRGKTINFDSTRGGISLVTEKGVHSSSRLLVQAARTTDAGAYQCAPDNAQSATARVHVLTGESPAAMQGSAHWADRFRLPTLIFFATIVYLTILIP
Summary
Uniprot
A0A212F1U5
A0A194QPN1
A0A194PP40
A0A194PHV5
A0A2A4J8J5
A0A212ER40
+ More
A0A194QN33 H9JLY3 A0A084W079 A0A182NR09 A0A0L7LGS4 A0A182PR62 A0A182FRY8 A0A182YNA9 A0A182QBM3 A0A182HK30 A0A182XNY4 Q7PUU3 Q176V6 A0A182VW88 A0A139WML4 A0A0R3P025 A0A1S4FCZ5 A0A1J1J020 A0A0R3P4Z1 A0A1B6E865 A0A1B6D7I4 A0A2S2Q445 A0A3B0KS10 A0A2S2NKY8 A0A1B6E3T8 A0A2H8TLG7 A0A151JTZ4 A0A154P4N4 A0A1B6CV50 A0A158NY86 V9IFQ7 B3MVT3 A0A088APM0 A0A3L8E428 A0A1J1IZP9 A0A151WMN9 A0A0Q9XNZ6 A0A0L0C4Y8 A0A1I8QAK9 E1ZYU6 A0A0M4EJH2 A0A232EDP6 A0A0Q9WKC4 Q9VY33 B3NV03 B4R4U3 B4IGD8 B4Q2T3 A0A1B6CPF3 A0A1W4VTS9 A0A1W4WTB8 A0A0L7QMS3 A0A336MND1 K7JB74 A0A0A1WYP1 A0A0Q9WNX0 A0A182TL83 A0A0R1EDW1 A0A1W4VUK0 A0A0L7R690 B4JL48 A0A182J9Y8 A0A151JXL8 A0A182L8A5 A0A182RG17 A0A336LZS2 K7JB75 A0A158NXR7 E0VMC5 A0A195CKU9 A0A1B6M443 A0A195ED84 A0A2S2QQD3 A0A1B6GTD3 A0A2S2PGS9 A0A2H8TZM1 A0A1B6KY95 J9K267 A0A1S4H0K5 A0A3L8E247 A0A182F3I4 A0A182IR75 A0A1B6M637 A0A1B6ENQ2 K7IXN4 A0A1Y1MRY7 A0A182PE72 A0A1B6CWP2 J9JRX7
A0A194QN33 H9JLY3 A0A084W079 A0A182NR09 A0A0L7LGS4 A0A182PR62 A0A182FRY8 A0A182YNA9 A0A182QBM3 A0A182HK30 A0A182XNY4 Q7PUU3 Q176V6 A0A182VW88 A0A139WML4 A0A0R3P025 A0A1S4FCZ5 A0A1J1J020 A0A0R3P4Z1 A0A1B6E865 A0A1B6D7I4 A0A2S2Q445 A0A3B0KS10 A0A2S2NKY8 A0A1B6E3T8 A0A2H8TLG7 A0A151JTZ4 A0A154P4N4 A0A1B6CV50 A0A158NY86 V9IFQ7 B3MVT3 A0A088APM0 A0A3L8E428 A0A1J1IZP9 A0A151WMN9 A0A0Q9XNZ6 A0A0L0C4Y8 A0A1I8QAK9 E1ZYU6 A0A0M4EJH2 A0A232EDP6 A0A0Q9WKC4 Q9VY33 B3NV03 B4R4U3 B4IGD8 B4Q2T3 A0A1B6CPF3 A0A1W4VTS9 A0A1W4WTB8 A0A0L7QMS3 A0A336MND1 K7JB74 A0A0A1WYP1 A0A0Q9WNX0 A0A182TL83 A0A0R1EDW1 A0A1W4VUK0 A0A0L7R690 B4JL48 A0A182J9Y8 A0A151JXL8 A0A182L8A5 A0A182RG17 A0A336LZS2 K7JB75 A0A158NXR7 E0VMC5 A0A195CKU9 A0A1B6M443 A0A195ED84 A0A2S2QQD3 A0A1B6GTD3 A0A2S2PGS9 A0A2H8TZM1 A0A1B6KY95 J9K267 A0A1S4H0K5 A0A3L8E247 A0A182F3I4 A0A182IR75 A0A1B6M637 A0A1B6ENQ2 K7IXN4 A0A1Y1MRY7 A0A182PE72 A0A1B6CWP2 J9JRX7
Pubmed
22118469
26354079
19121390
24438588
26227816
25244985
+ More
12364791 14747013 17210077 17510324 18362917 19820115 15632085 17994087 21347285 30249741 26108605 20798317 28648823 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20075255 25830018 20966253 20566863 28004739
12364791 14747013 17210077 17510324 18362917 19820115 15632085 17994087 21347285 30249741 26108605 20798317 28648823 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20075255 25830018 20966253 20566863 28004739
EMBL
AGBW02010791
OWR47706.1
KQ461194
KPJ06935.1
KQ459603
KPI92895.1
+ More
KPI92897.1 NWSH01002656 PCG67732.1 AGBW02013172 OWR43914.1 KPJ06933.1 BABH01005427 BABH01005428 ATLV01019094 KE525262 KFB43623.1 JTDY01001184 KOB74620.1 AXCN02001484 AXCN02001485 APCN01000815 AAAB01008987 EAA00989.5 CH477382 EAT42180.1 KQ971312 KYB29210.1 CH379064 KRT06622.1 CVRI01000065 CRL05743.1 KRT06621.1 GEDC01012553 GEDC01003178 JAS24745.1 JAS34120.1 GEDC01015655 JAS21643.1 GGMS01003322 MBY72525.1 OUUW01000015 SPP88696.1 GGMR01005205 MBY17824.1 GEDC01004724 JAS32574.1 GFXV01003191 MBW14996.1 KQ981799 KYN35656.1 KQ434814 KZC06827.1 GEDC01020047 JAS17251.1 ADTU01003391 ADTU01003392 ADTU01003393 ADTU01003394 ADTU01003395 ADTU01003396 ADTU01003397 ADTU01003398 ADTU01003399 ADTU01003400 JR041747 AEY59502.1 CH902625 EDV35078.1 QOIP01000001 RLU27302.1 CRL05739.1 KQ982934 KYQ49126.1 CH933813 KRG07402.1 JRES01000902 KNC27443.1 GL435242 EFN73636.1 CP012528 ALC49291.1 NNAY01006059 OXU16465.1 CH940651 KRF82033.1 KRF82034.1 KRF82036.1 KRF82038.1 KRF82039.1 KRF82040.1 AE014298 AY060637 AAF48372.2 AAL28185.1 AHN59709.1 CH954180 EDV47101.1 CM000366 EDX17900.1 CH480835 EDW48872.1 CM000162 EDX01677.1 KRK06287.1 KRK06288.1 KRK06289.1 KRK06290.1 GEDC01021959 JAS15339.1 KQ414868 KOC59947.1 UFQT01001458 SSX30649.1 GBXI01010536 GBXI01005761 GBXI01002155 JAD03756.1 JAD08531.1 JAD12137.1 KRF82032.1 KRK06291.1 KQ414648 KOC66353.1 CH916370 EDW00301.1 KQ981567 KYN39797.1 UFQT01000229 SSX22073.1 ADTU01003373 ADTU01003374 ADTU01003375 ADTU01003376 ADTU01003377 ADTU01003378 ADTU01003379 ADTU01003380 DS235306 EEB14531.1 KQ977622 KYN01333.1 GEBQ01009282 JAT30695.1 KQ979074 KYN23061.1 GGMS01010680 MBY79883.1 GECZ01004098 JAS65671.1 GGMR01016031 MBY28650.1 GFXV01007892 MBW19697.1 GEBQ01023602 JAT16375.1 ABLF02023164 ABLF02023168 ABLF02023170 ABLF02023171 ABLF02023172 ABLF02028992 ABLF02028994 ABLF02028999 AAAB01008984 RLU26716.1 GEBQ01008578 JAT31399.1 GECZ01030217 GECZ01002715 JAS39552.1 JAS67054.1 AAZX01000015 AAZX01000039 AAZX01005416 AAZX01007570 AAZX01007787 AAZX01009562 AAZX01012879 AAZX01013757 AAZX01022366 GEZM01026071 JAV87260.1 GEDC01019431 JAS17867.1 ABLF02039192 ABLF02039203 ABLF02039205 ABLF02039207 ABLF02039208 ABLF02039696 ABLF02039705 ABLF02039706 ABLF02053971 ABLF02064527
KPI92897.1 NWSH01002656 PCG67732.1 AGBW02013172 OWR43914.1 KPJ06933.1 BABH01005427 BABH01005428 ATLV01019094 KE525262 KFB43623.1 JTDY01001184 KOB74620.1 AXCN02001484 AXCN02001485 APCN01000815 AAAB01008987 EAA00989.5 CH477382 EAT42180.1 KQ971312 KYB29210.1 CH379064 KRT06622.1 CVRI01000065 CRL05743.1 KRT06621.1 GEDC01012553 GEDC01003178 JAS24745.1 JAS34120.1 GEDC01015655 JAS21643.1 GGMS01003322 MBY72525.1 OUUW01000015 SPP88696.1 GGMR01005205 MBY17824.1 GEDC01004724 JAS32574.1 GFXV01003191 MBW14996.1 KQ981799 KYN35656.1 KQ434814 KZC06827.1 GEDC01020047 JAS17251.1 ADTU01003391 ADTU01003392 ADTU01003393 ADTU01003394 ADTU01003395 ADTU01003396 ADTU01003397 ADTU01003398 ADTU01003399 ADTU01003400 JR041747 AEY59502.1 CH902625 EDV35078.1 QOIP01000001 RLU27302.1 CRL05739.1 KQ982934 KYQ49126.1 CH933813 KRG07402.1 JRES01000902 KNC27443.1 GL435242 EFN73636.1 CP012528 ALC49291.1 NNAY01006059 OXU16465.1 CH940651 KRF82033.1 KRF82034.1 KRF82036.1 KRF82038.1 KRF82039.1 KRF82040.1 AE014298 AY060637 AAF48372.2 AAL28185.1 AHN59709.1 CH954180 EDV47101.1 CM000366 EDX17900.1 CH480835 EDW48872.1 CM000162 EDX01677.1 KRK06287.1 KRK06288.1 KRK06289.1 KRK06290.1 GEDC01021959 JAS15339.1 KQ414868 KOC59947.1 UFQT01001458 SSX30649.1 GBXI01010536 GBXI01005761 GBXI01002155 JAD03756.1 JAD08531.1 JAD12137.1 KRF82032.1 KRK06291.1 KQ414648 KOC66353.1 CH916370 EDW00301.1 KQ981567 KYN39797.1 UFQT01000229 SSX22073.1 ADTU01003373 ADTU01003374 ADTU01003375 ADTU01003376 ADTU01003377 ADTU01003378 ADTU01003379 ADTU01003380 DS235306 EEB14531.1 KQ977622 KYN01333.1 GEBQ01009282 JAT30695.1 KQ979074 KYN23061.1 GGMS01010680 MBY79883.1 GECZ01004098 JAS65671.1 GGMR01016031 MBY28650.1 GFXV01007892 MBW19697.1 GEBQ01023602 JAT16375.1 ABLF02023164 ABLF02023168 ABLF02023170 ABLF02023171 ABLF02023172 ABLF02028992 ABLF02028994 ABLF02028999 AAAB01008984 RLU26716.1 GEBQ01008578 JAT31399.1 GECZ01030217 GECZ01002715 JAS39552.1 JAS67054.1 AAZX01000015 AAZX01000039 AAZX01005416 AAZX01007570 AAZX01007787 AAZX01009562 AAZX01012879 AAZX01013757 AAZX01022366 GEZM01026071 JAV87260.1 GEDC01019431 JAS17867.1 ABLF02039192 ABLF02039203 ABLF02039205 ABLF02039207 ABLF02039208 ABLF02039696 ABLF02039705 ABLF02039706 ABLF02053971 ABLF02064527
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000005204
UP000030765
+ More
UP000075884 UP000037510 UP000075885 UP000069272 UP000076408 UP000075886 UP000075840 UP000076407 UP000007062 UP000008820 UP000075920 UP000007266 UP000001819 UP000183832 UP000268350 UP000078541 UP000076502 UP000005205 UP000007801 UP000005203 UP000279307 UP000075809 UP000009192 UP000037069 UP000095300 UP000000311 UP000092553 UP000215335 UP000008792 UP000000803 UP000008711 UP000000304 UP000001292 UP000002282 UP000192221 UP000192223 UP000053825 UP000002358 UP000075902 UP000001070 UP000075880 UP000075882 UP000075900 UP000009046 UP000078542 UP000078492 UP000007819
UP000075884 UP000037510 UP000075885 UP000069272 UP000076408 UP000075886 UP000075840 UP000076407 UP000007062 UP000008820 UP000075920 UP000007266 UP000001819 UP000183832 UP000268350 UP000078541 UP000076502 UP000005205 UP000007801 UP000005203 UP000279307 UP000075809 UP000009192 UP000037069 UP000095300 UP000000311 UP000092553 UP000215335 UP000008792 UP000000803 UP000008711 UP000000304 UP000001292 UP000002282 UP000192221 UP000192223 UP000053825 UP000002358 UP000075902 UP000001070 UP000075880 UP000075882 UP000075900 UP000009046 UP000078542 UP000078492 UP000007819
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A212F1U5
A0A194QPN1
A0A194PP40
A0A194PHV5
A0A2A4J8J5
A0A212ER40
+ More
A0A194QN33 H9JLY3 A0A084W079 A0A182NR09 A0A0L7LGS4 A0A182PR62 A0A182FRY8 A0A182YNA9 A0A182QBM3 A0A182HK30 A0A182XNY4 Q7PUU3 Q176V6 A0A182VW88 A0A139WML4 A0A0R3P025 A0A1S4FCZ5 A0A1J1J020 A0A0R3P4Z1 A0A1B6E865 A0A1B6D7I4 A0A2S2Q445 A0A3B0KS10 A0A2S2NKY8 A0A1B6E3T8 A0A2H8TLG7 A0A151JTZ4 A0A154P4N4 A0A1B6CV50 A0A158NY86 V9IFQ7 B3MVT3 A0A088APM0 A0A3L8E428 A0A1J1IZP9 A0A151WMN9 A0A0Q9XNZ6 A0A0L0C4Y8 A0A1I8QAK9 E1ZYU6 A0A0M4EJH2 A0A232EDP6 A0A0Q9WKC4 Q9VY33 B3NV03 B4R4U3 B4IGD8 B4Q2T3 A0A1B6CPF3 A0A1W4VTS9 A0A1W4WTB8 A0A0L7QMS3 A0A336MND1 K7JB74 A0A0A1WYP1 A0A0Q9WNX0 A0A182TL83 A0A0R1EDW1 A0A1W4VUK0 A0A0L7R690 B4JL48 A0A182J9Y8 A0A151JXL8 A0A182L8A5 A0A182RG17 A0A336LZS2 K7JB75 A0A158NXR7 E0VMC5 A0A195CKU9 A0A1B6M443 A0A195ED84 A0A2S2QQD3 A0A1B6GTD3 A0A2S2PGS9 A0A2H8TZM1 A0A1B6KY95 J9K267 A0A1S4H0K5 A0A3L8E247 A0A182F3I4 A0A182IR75 A0A1B6M637 A0A1B6ENQ2 K7IXN4 A0A1Y1MRY7 A0A182PE72 A0A1B6CWP2 J9JRX7
A0A194QN33 H9JLY3 A0A084W079 A0A182NR09 A0A0L7LGS4 A0A182PR62 A0A182FRY8 A0A182YNA9 A0A182QBM3 A0A182HK30 A0A182XNY4 Q7PUU3 Q176V6 A0A182VW88 A0A139WML4 A0A0R3P025 A0A1S4FCZ5 A0A1J1J020 A0A0R3P4Z1 A0A1B6E865 A0A1B6D7I4 A0A2S2Q445 A0A3B0KS10 A0A2S2NKY8 A0A1B6E3T8 A0A2H8TLG7 A0A151JTZ4 A0A154P4N4 A0A1B6CV50 A0A158NY86 V9IFQ7 B3MVT3 A0A088APM0 A0A3L8E428 A0A1J1IZP9 A0A151WMN9 A0A0Q9XNZ6 A0A0L0C4Y8 A0A1I8QAK9 E1ZYU6 A0A0M4EJH2 A0A232EDP6 A0A0Q9WKC4 Q9VY33 B3NV03 B4R4U3 B4IGD8 B4Q2T3 A0A1B6CPF3 A0A1W4VTS9 A0A1W4WTB8 A0A0L7QMS3 A0A336MND1 K7JB74 A0A0A1WYP1 A0A0Q9WNX0 A0A182TL83 A0A0R1EDW1 A0A1W4VUK0 A0A0L7R690 B4JL48 A0A182J9Y8 A0A151JXL8 A0A182L8A5 A0A182RG17 A0A336LZS2 K7JB75 A0A158NXR7 E0VMC5 A0A195CKU9 A0A1B6M443 A0A195ED84 A0A2S2QQD3 A0A1B6GTD3 A0A2S2PGS9 A0A2H8TZM1 A0A1B6KY95 J9K267 A0A1S4H0K5 A0A3L8E247 A0A182F3I4 A0A182IR75 A0A1B6M637 A0A1B6ENQ2 K7IXN4 A0A1Y1MRY7 A0A182PE72 A0A1B6CWP2 J9JRX7
PDB
6EG0
E-value=2.09724e-47,
Score=475
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.693357
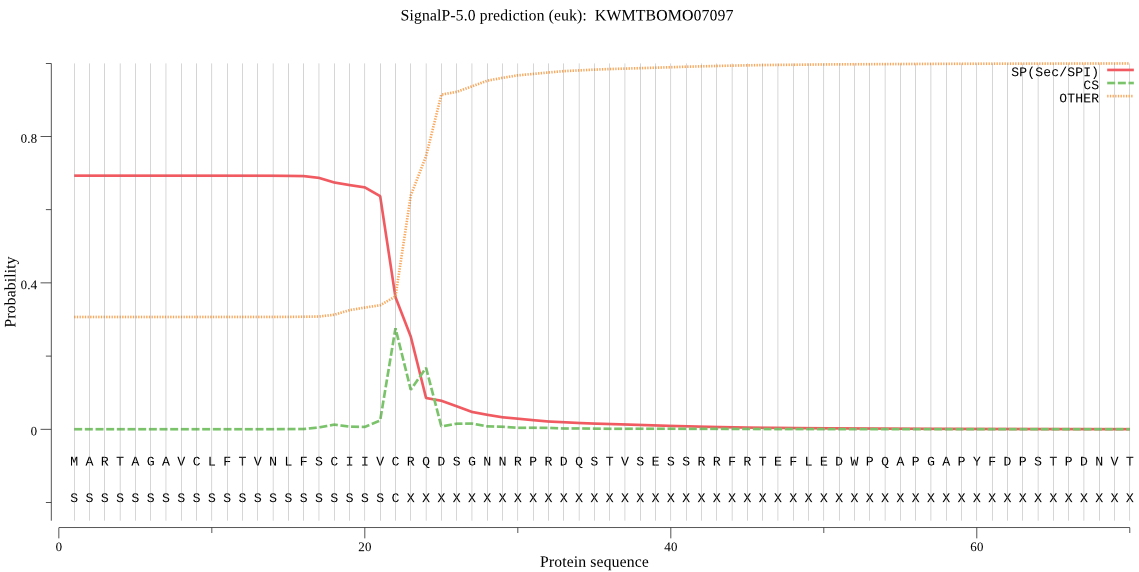
Length:
295
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
29.191
Exp number, first 60 AAs:
12.60219
Total prob of N-in:
0.16496
POSSIBLE N-term signal
sequence
outside
1 - 295

Population Genetic Test Statistics
Pi
183.539327
Theta
176.428058
Tajima's D
0.179107
CLR
0.526072
CSRT
0.422528873556322
Interpretation
Uncertain
