Gene
KWMTBOMO07095
Pre Gene Modal
BGIBMGA004264
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_4-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.586
Sequence
CDS
ATGACTGAAACATCCACAACAAACATGGTGTCAGGTGTGGGGTTCCTGCCACTTTTACCCAGTATGGACTTCAAAGGTAACCTTGCCGAAAATTTCAAAATGTATCGTCAAAGGTTGGAGATATATTTGTCTGCTAATGGCCTTGATGACGCCAAAGACAAAAGAAAAATATCAATTCTGCTGAATTTTATTGGCGAAAACGGAATCAAAATCTACAATTCTTTCAACTTAAACGCATCTGAATCACATACTTTTGAACACGTGCTCAAAAAGTTTGAAGCGTACTTTGAACCAAAGAAAAGTCTAACTATAGCAAGGAACGAGTTTTTTACTGTTACACAAGATGAAGACAGGTTGGAGGATTACTTTCAAAGAGTAATAAATCTCGGTATTCAATGCGAACTTGGAGATCTCAGAGAAGGGTTGACAGTGGGAAAGATAATTTCAGGGTTAGACTCCAAATATAATAATCTCAAAACACGATTAATGTCGGAAGACGATAGTAAATTAACATTAGATTATGTTATGAAATACTTACTGAGTGCCGAAACCTCTAGAAAATATATAGAAGAAAAAACAAATTCAAAGGACAGCGATGAGTCCTCCATCCTATATGTTAATAAAAAATCAATAAGGAACTGTAGAAACTGTGGCACATCTCACAATATAAATAAATGCCCAGCATACGGCAAAGAGTGTCTGAAGTGCAAGAAACTTAACCATTTTAGTAAAGTATGCAGATCAGCCAAGAAACCAAATTTTTATCAAAAGCAAGTCAATGTCCTTGAGGATCCAGATAAGAACAAAGAGGATGAAGCTGAAGGAGATCTCTTTGTTGGTTATGCAGGCAATGAAATAATTCAAGATTTTATAGTCAATAACATCAAAATTCCCATCAAAATGGACACTGGAGCTGCATGCAACGTGATTGGGAGCGAAGTATGTAAATTAATTAATATAAAAACCTCAGAAATTTTGCCCTGTGACAAAAAGGTCATTCAACTAGATGGAAAATGTGTACCAGTCAGGGGTACTTGTTGGTTGAATATTATTCATCCTAATAGCAAAAAATACACTGTTAAGTTCATCATAATAGAAGGAAATATTCCTACTTTACTCGGGTGCAGAACATGTTTAACTTATAATTTTATTTCAGCAAACACTAACCTTGTTAACATACATAATGTGACAGTTGGAAAAGACACAACATTCAATTTAATAAAAAAATTAAAAAATGATTTCAGTGAGCTCTTCAGTGGAGGTCTTGGTTGCATTCCTGGCACAGTAAACATTCAGCTAAGTAGTGAAGCAGTCCCTACAGTGCAGGCTGCTAGACGTGTTCCATTTGCATTAATGCCTGAACTGCGCCGAGAATTAGATCATATGGAAGCAATAGGGGTTATTGAAAAGGTAGATAAGCCCACCAAATGGGTAAATAATATAGTGCCTGTTAGAAAAACGAATGATGATATACCTGATATTGTATTAGATAATAGCATGTGTCAGCCTTCAGCTAAATATGTTATACAAGAGCCTAGCTTAGAAAAAATAGTATGTGGAAGTAACATGGCAACCCAGGCGAGTCCAGCAAGATCACGGGAGAGAAGGGAGCAGAGAATACTTCAAAGCGTCTCCGGAGAGTTTCGTTCAGGAGAGCTGACATGTATACTAGGACCTTCGGGGGCTGGAAAGTCTACTTTACTCAACATCTTAGCAGGATACACGTTGAACGGCGTCACCGGACGTATATCAGTGAACGGACACGTGCGAGACATGAGGGTCTTCAAGAAGCTGAGCTCGTACATCATGCAGGATGATCTGCTGCAGCCAAGACTGACCGTGCAAGAGTCCATGTGGATCGCTGCTGATCTGAAACTCGGCAAGGAACTGGGAAAGCCGGAGAAGGAACTCATTATTGAAGAAATTCTACAAACACTAGGTCTGTGGGATCACAGAGACACAAGGTCTGAAAAACTGTCTGGAGGACAATCGAAACGTCTCACTGTCGCCCTGGAGCTGGTCAACAATCCACCCATCATATTTCTGGATGAGCCTACCACGTGCGTATACTCTTCCACAATTACTGTATAG
Protein
MTETSTTNMVSGVGFLPLLPSMDFKGNLAENFKMYRQRLEIYLSANGLDDAKDKRKISILLNFIGENGIKIYNSFNLNASESHTFEHVLKKFEAYFEPKKSLTIARNEFFTVTQDEDRLEDYFQRVINLGIQCELGDLREGLTVGKIISGLDSKYNNLKTRLMSEDDSKLTLDYVMKYLLSAETSRKYIEEKTNSKDSDESSILYVNKKSIRNCRNCGTSHNINKCPAYGKECLKCKKLNHFSKVCRSAKKPNFYQKQVNVLEDPDKNKEDEAEGDLFVGYAGNEIIQDFIVNNIKIPIKMDTGAACNVIGSEVCKLINIKTSEILPCDKKVIQLDGKCVPVRGTCWLNIIHPNSKKYTVKFIIIEGNIPTLLGCRTCLTYNFISANTNLVNIHNVTVGKDTTFNLIKKLKNDFSELFSGGLGCIPGTVNIQLSSEAVPTVQAARRVPFALMPELRRELDHMEAIGVIEKVDKPTKWVNNIVPVRKTNDDIPDIVLDNSMCQPSAKYVIQEPSLEKIVCGSNMATQASPARSRERREQRILQSVSGEFRSGELTCILGPSGAGKSTLLNILAGYTLNGVTGRISVNGHVRDMRVFKKLSSYIMQDDLLQPRLTVQESMWIAADLKLGKELGKPEKELIIEEILQTLGLWDHRDTRSEKLSGGQSKRLTVALELVNNPPIIFLDEPTTCVYSSTITV
Summary
Uniprot
A0A2G8L9H5
A0A3P8S414
A0A3B1JKR1
A0A3P8Q6C6
A0A2G8KKG5
A0A1A8UGA0
+ More
A0A3P9M8I3 A0A2S2N8I3 A0A3B3HLX7 X1WTY2 A0A069DZS8 A0A069DY08 A0A146SN99 A0A3Q0JFZ1 A0A146TAZ6 A0A3P9HXP1 A0A1Y1MUX5 A0A1Y1MCJ7 A0A2G8LMB1 A0A0A9Z4Y4 L7LWW6 A0A023EY30 A0A3P8RKT7 A0A146LC45 A0A1Y1M671 A0A1S3ICH8 A0A3P8RHF8 A0A3P9AYU2 A0A2G8KHQ8 A0A3P8PD24 A0A1S3HTD1 A0A075EIL5 A0A1S3DNR0 A0A1S3DM37 A0A1S3DH71 A0A1Y1JV53 A0A2G8KFU3 A0A3B3I1S4 A0A0J7K756 A0A2S2NI46 A0A3P9LZF9 A0A3P9K9H2 A0A1Y1NHV7 A0A293LIZ7 A0A3B3C0E1 W4YTX1 A0A2G8K365 A0A1Y1K861 A0A147BN28 A0A3Q0J4U3 A0A3P8QAD1 A0A1Y1NDC5 I3KYE2 A0A3B3H7E4 A0A1A8JWC8 W4YGV7 J9LDY4 A0A1Y1K7Z4 A0A1A8QDB9 A0A2B4S589 W4Y1E4 A0A2B4S229 A0A1A8QB08 A0A1A8N283 A0A1A8DQM9 A0A1A8P545
A0A3P9M8I3 A0A2S2N8I3 A0A3B3HLX7 X1WTY2 A0A069DZS8 A0A069DY08 A0A146SN99 A0A3Q0JFZ1 A0A146TAZ6 A0A3P9HXP1 A0A1Y1MUX5 A0A1Y1MCJ7 A0A2G8LMB1 A0A0A9Z4Y4 L7LWW6 A0A023EY30 A0A3P8RKT7 A0A146LC45 A0A1Y1M671 A0A1S3ICH8 A0A3P8RHF8 A0A3P9AYU2 A0A2G8KHQ8 A0A3P8PD24 A0A1S3HTD1 A0A075EIL5 A0A1S3DNR0 A0A1S3DM37 A0A1S3DH71 A0A1Y1JV53 A0A2G8KFU3 A0A3B3I1S4 A0A0J7K756 A0A2S2NI46 A0A3P9LZF9 A0A3P9K9H2 A0A1Y1NHV7 A0A293LIZ7 A0A3B3C0E1 W4YTX1 A0A2G8K365 A0A1Y1K861 A0A147BN28 A0A3Q0J4U3 A0A3P8QAD1 A0A1Y1NDC5 I3KYE2 A0A3B3H7E4 A0A1A8JWC8 W4YGV7 J9LDY4 A0A1Y1K7Z4 A0A1A8QDB9 A0A2B4S589 W4Y1E4 A0A2B4S229 A0A1A8QB08 A0A1A8N283 A0A1A8DQM9 A0A1A8P545
Pubmed
EMBL
MRZV01000161
PIK56894.1
MRZV01000519
PIK48493.1
HADY01023773
HAEJ01006887
+ More
SBS47344.1 GGMR01000868 MBY13487.1 ABLF02013930 ABLF02013934 GBGD01000115 JAC88774.1 GBGD01000119 JAC88770.1 GCES01104253 JAQ82069.1 GCES01096219 JAQ90103.1 GEZM01020000 JAV89472.1 GEZM01035137 JAV83383.1 MRZV01000033 PIK61397.1 GBHO01004090 JAG39514.1 GACK01009641 JAA55393.1 GBBI01004745 JAC13967.1 GDHC01013957 JAQ04672.1 GEZM01041944 JAV80090.1 MRZV01000576 PIK47528.1 KF319019 AIE48224.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1 MRZV01000618 PIK46835.1 LBMM01012476 KMQ86207.1 GGMR01004179 MBY16798.1 GEZM01001973 JAV97431.1 GFWV01003962 MAA28692.1 AAGJ04045137 MRZV01000935 PIK42441.1 GEZM01089256 JAV57584.1 GEGO01003204 JAR92200.1 GEZM01011167 JAV93617.1 AERX01070080 HAEE01004368 SBR24388.1 AAGJ04164197 ABLF02010707 GEZM01089257 JAV57582.1 HAEI01005002 SBR91338.1 LSMT01000199 PFX23692.1 AAGJ04066141 LSMT01000218 PFX23103.1 HAEG01011680 SBR90379.1 HAEF01021833 SBR62992.1 HADZ01002490 HAEA01007842 SBQ36322.1 HAEG01006212 SBR76132.1
SBS47344.1 GGMR01000868 MBY13487.1 ABLF02013930 ABLF02013934 GBGD01000115 JAC88774.1 GBGD01000119 JAC88770.1 GCES01104253 JAQ82069.1 GCES01096219 JAQ90103.1 GEZM01020000 JAV89472.1 GEZM01035137 JAV83383.1 MRZV01000033 PIK61397.1 GBHO01004090 JAG39514.1 GACK01009641 JAA55393.1 GBBI01004745 JAC13967.1 GDHC01013957 JAQ04672.1 GEZM01041944 JAV80090.1 MRZV01000576 PIK47528.1 KF319019 AIE48224.1 GEZM01102055 GEZM01102054 GEZM01102053 GEZM01102051 JAV52011.1 MRZV01000618 PIK46835.1 LBMM01012476 KMQ86207.1 GGMR01004179 MBY16798.1 GEZM01001973 JAV97431.1 GFWV01003962 MAA28692.1 AAGJ04045137 MRZV01000935 PIK42441.1 GEZM01089256 JAV57584.1 GEGO01003204 JAR92200.1 GEZM01011167 JAV93617.1 AERX01070080 HAEE01004368 SBR24388.1 AAGJ04164197 ABLF02010707 GEZM01089257 JAV57582.1 HAEI01005002 SBR91338.1 LSMT01000199 PFX23692.1 AAGJ04066141 LSMT01000218 PFX23103.1 HAEG01011680 SBR90379.1 HAEF01021833 SBR62992.1 HADZ01002490 HAEA01007842 SBQ36322.1 HAEG01006212 SBR76132.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2G8L9H5
A0A3P8S414
A0A3B1JKR1
A0A3P8Q6C6
A0A2G8KKG5
A0A1A8UGA0
+ More
A0A3P9M8I3 A0A2S2N8I3 A0A3B3HLX7 X1WTY2 A0A069DZS8 A0A069DY08 A0A146SN99 A0A3Q0JFZ1 A0A146TAZ6 A0A3P9HXP1 A0A1Y1MUX5 A0A1Y1MCJ7 A0A2G8LMB1 A0A0A9Z4Y4 L7LWW6 A0A023EY30 A0A3P8RKT7 A0A146LC45 A0A1Y1M671 A0A1S3ICH8 A0A3P8RHF8 A0A3P9AYU2 A0A2G8KHQ8 A0A3P8PD24 A0A1S3HTD1 A0A075EIL5 A0A1S3DNR0 A0A1S3DM37 A0A1S3DH71 A0A1Y1JV53 A0A2G8KFU3 A0A3B3I1S4 A0A0J7K756 A0A2S2NI46 A0A3P9LZF9 A0A3P9K9H2 A0A1Y1NHV7 A0A293LIZ7 A0A3B3C0E1 W4YTX1 A0A2G8K365 A0A1Y1K861 A0A147BN28 A0A3Q0J4U3 A0A3P8QAD1 A0A1Y1NDC5 I3KYE2 A0A3B3H7E4 A0A1A8JWC8 W4YGV7 J9LDY4 A0A1Y1K7Z4 A0A1A8QDB9 A0A2B4S589 W4Y1E4 A0A2B4S229 A0A1A8QB08 A0A1A8N283 A0A1A8DQM9 A0A1A8P545
A0A3P9M8I3 A0A2S2N8I3 A0A3B3HLX7 X1WTY2 A0A069DZS8 A0A069DY08 A0A146SN99 A0A3Q0JFZ1 A0A146TAZ6 A0A3P9HXP1 A0A1Y1MUX5 A0A1Y1MCJ7 A0A2G8LMB1 A0A0A9Z4Y4 L7LWW6 A0A023EY30 A0A3P8RKT7 A0A146LC45 A0A1Y1M671 A0A1S3ICH8 A0A3P8RHF8 A0A3P9AYU2 A0A2G8KHQ8 A0A3P8PD24 A0A1S3HTD1 A0A075EIL5 A0A1S3DNR0 A0A1S3DM37 A0A1S3DH71 A0A1Y1JV53 A0A2G8KFU3 A0A3B3I1S4 A0A0J7K756 A0A2S2NI46 A0A3P9LZF9 A0A3P9K9H2 A0A1Y1NHV7 A0A293LIZ7 A0A3B3C0E1 W4YTX1 A0A2G8K365 A0A1Y1K861 A0A147BN28 A0A3Q0J4U3 A0A3P8QAD1 A0A1Y1NDC5 I3KYE2 A0A3B3H7E4 A0A1A8JWC8 W4YGV7 J9LDY4 A0A1Y1K7Z4 A0A1A8QDB9 A0A2B4S589 W4Y1E4 A0A2B4S229 A0A1A8QB08 A0A1A8N283 A0A1A8DQM9 A0A1A8P545
PDB
6FFC
E-value=1.60497e-16,
Score=213
Ontologies
GO
Topology
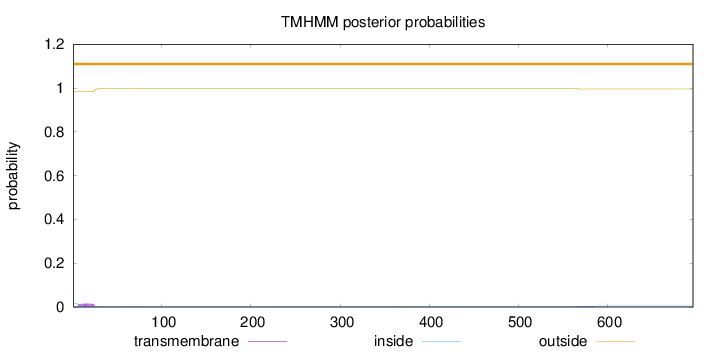
Length:
696
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.38968
Exp number, first 60 AAs:
0.28581
Total prob of N-in:
0.01649
outside
1 - 696
Population Genetic Test Statistics
Pi
259.154569
Theta
167.644185
Tajima's D
1.875419
CLR
0.02267
CSRT
0.859907004649768
Interpretation
Uncertain
