Gene
KWMTBOMO07094
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_4-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.182
Sequence
CDS
ATGGAAGTGCACCTACTGCGTCGGGAATACTTCAATCGCTGGTACAGCTTAAAGGCTTATTATGCTGCGCTGAGTTTTGCCACTATGCCTGCGATGACTTTACTTGGCCTCATATTCCTGGTTATAACGTATTTCATGTCAGGGCAGCTGCTAGAGTGGGAGAGGTTTACATTGTTCTCTATCAGCGGACTGCTCACGGGAATATGTTCCGAGGGCTTAGGTCTCGCCATAGGCGCAGCACTGAGTGTTACTAACGGCTCTATCGTCGGACCATCGATAGTGTCACCACTCCTGGTTCTCTGCTGCTACGGAATGGGTTTCGGTCCTTACATAGAGGGCACCATGCGCGCCCTAATGTCCGCCTCGTATCTGCGTTACGGTCTTTCCGGATTAACTTTATCCCTAGAGTCCGTAGTGTTTTACCGCATTTTGGATTTGAACAGCATGAAAGCTCATATACTGCACAACATGCACCAACCTAAGATTATAGAAAGGGGAGATTTCCTAAAGACTAGCTCGTTCGCTAGTTCTCCCATGGGGGACCAGACTACGCGTTTCTGA
Protein
MEVHLLRREYFNRWYSLKAYYAALSFATMPAMTLLGLIFLVITYFMSGQLLEWERFTLFSISGLLTGICSEGLGLAIGAALSVTNGSIVGPSIVSPLLVLCCYGMGFGPYIEGTMRALMSASYLRYGLSGLTLSLESVVFYRILDLNSMKAHILHNMHQPKIIERGDFLKTSSFASSPMGDQTTRF
Summary
Uniprot
A0A1D9C109
S4NIB2
A0A194PHU9
A0A194PIL7
A0A2A4JSF5
A0A2H1WKT2
+ More
A0A212EYH0 N6UL09 N6TPJ2 A0A139WK45 U4UVY2 J3JWW1 A0A2R7WJI2 A0A2J7PSR5 A0A2J7R9W6 T1I3H7 A0A023F2D5 A0A0P4VIF7 A0A2R4FXE8 A0A0U9HZ98 A0A067R385 A0A067QKC2 A0A2P8YSB2 Q8MS57 Q9VTL3 Q8SXX6 A0A0J9RTT3 B4HEW3 B3NGY3 B4PFL8 A0A1W4VU95 B4HAI3 A0A1B6E8X1 A0A158V6Y6 B4KUR3 B3M4A1 A0A1B6DIR7 Q2LZ69 A0A3B0JZT0 A0A3B0JVM2 A0A1S4EI61 B4N0M2 B4N3B8 A0A3B0K6S5 B4GQS2 B4KFN7 A0A146M787 B4MDB5 A0A1B6CDE3 A0A1B6EE46 A0A1B6I4E0 J9JXC1 B4LCP8 A0A1W4X256 A0A0K8V5S5 A0A0M4EG47 A0A0T6ATZ0 W8C2Y8 A0A0A9YE03 A0A1B6DT64 A0A2P8Z045 A0A1B6DAM8 A0A2S2NDX0 B4J2J5 A0A2S2RAU7 B4JD50 A0A3R5USX7 A0A2S2QQM8 W8AXI7 A0A1B6G7Z6 A0A0A1XNQ2 W8B4R5 Q16NT8 A0A158V4U5 A0A034WRH9 A0A2H4TES4 A0A0K8VTK0 A0A1E1W184 B0WLD9 A0A0L0CDT2 A0A1Q3FNT7 J9JSX9 A0A1Q3FNC5 A0A1B6LXW2 A0A1Q3FNE6 A0A0M3QTZ8 A0A0K8WFN7 A0A034V8F4 A0A158V1B5 A0A0A1WY89 A0A1W4UQ15 A0A2S2PAR8 A0A2H4TES0 A0A2H8TW86 A0A0L7LFT1 A0A1B6DFY5 J9K4G9 A0A2S2QS91
A0A212EYH0 N6UL09 N6TPJ2 A0A139WK45 U4UVY2 J3JWW1 A0A2R7WJI2 A0A2J7PSR5 A0A2J7R9W6 T1I3H7 A0A023F2D5 A0A0P4VIF7 A0A2R4FXE8 A0A0U9HZ98 A0A067R385 A0A067QKC2 A0A2P8YSB2 Q8MS57 Q9VTL3 Q8SXX6 A0A0J9RTT3 B4HEW3 B3NGY3 B4PFL8 A0A1W4VU95 B4HAI3 A0A1B6E8X1 A0A158V6Y6 B4KUR3 B3M4A1 A0A1B6DIR7 Q2LZ69 A0A3B0JZT0 A0A3B0JVM2 A0A1S4EI61 B4N0M2 B4N3B8 A0A3B0K6S5 B4GQS2 B4KFN7 A0A146M787 B4MDB5 A0A1B6CDE3 A0A1B6EE46 A0A1B6I4E0 J9JXC1 B4LCP8 A0A1W4X256 A0A0K8V5S5 A0A0M4EG47 A0A0T6ATZ0 W8C2Y8 A0A0A9YE03 A0A1B6DT64 A0A2P8Z045 A0A1B6DAM8 A0A2S2NDX0 B4J2J5 A0A2S2RAU7 B4JD50 A0A3R5USX7 A0A2S2QQM8 W8AXI7 A0A1B6G7Z6 A0A0A1XNQ2 W8B4R5 Q16NT8 A0A158V4U5 A0A034WRH9 A0A2H4TES4 A0A0K8VTK0 A0A1E1W184 B0WLD9 A0A0L0CDT2 A0A1Q3FNT7 J9JSX9 A0A1Q3FNC5 A0A1B6LXW2 A0A1Q3FNE6 A0A0M3QTZ8 A0A0K8WFN7 A0A034V8F4 A0A158V1B5 A0A0A1WY89 A0A1W4UQ15 A0A2S2PAR8 A0A2H4TES0 A0A2H8TW86 A0A0L7LFT1 A0A1B6DFY5 J9K4G9 A0A2S2QS91
Pubmed
23622113
26354079
22118469
23537049
18362917
19820115
+ More
22516182 25474469 27129103 24845553 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17994087 17550304 18057021 15632085 26823975 24495485 25401762 25830018 17510324 25348373 29121518 26108605 26227816
22516182 25474469 27129103 24845553 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17994087 17550304 18057021 15632085 26823975 24495485 25401762 25830018 17510324 25348373 29121518 26108605 26227816
EMBL
KX793137
AOY08602.1
GAIX01014139
JAA78421.1
KQ459603
KPI92892.1
+ More
KPI92893.1 NWSH01000663 PCG74941.1 ODYU01009275 SOQ53576.1 AGBW02011531 OWR46507.1 APGK01013174 KB738930 ENN82435.1 APGK01013159 APGK01013160 KB738929 ENN82439.1 KQ971338 KYB28165.1 KB632390 ERL94411.1 BT127729 AEE62691.1 KK854769 PTY18555.1 NEVH01021923 PNF19374.1 NEVH01006580 PNF37625.1 ACPB03010505 GBBI01003272 JAC15440.1 GDKW01002928 JAI53667.1 MF034805 AVT42171.1 GARF01000042 JAC88905.1 KK853032 KDR12258.1 KK853336 KDR08336.1 PYGN01000390 PSN47138.1 AY119083 AAM50943.1 AE014296 BT056236 AAF50035.2 ACL68683.1 AY075521 AAL68329.1 CM002912 KMY99032.1 CH480815 EDW41132.1 CH954178 EDV51440.1 CM000159 EDW94167.2 CH479240 EDW37581.1 GEDC01002932 JAS34366.1 KF828794 AIN44111.1 CH933809 EDW19319.1 CH902618 EDV39371.1 KPU78026.1 GEDC01011746 JAS25552.1 CH379069 EAL29640.1 OUUW01000004 SPP79199.1 OUUW01000002 SPP76781.1 CH963920 EDW77635.2 CH964062 EDW78857.2 SPP79198.1 CH479187 EDW39944.1 CH933807 EDW11002.1 KRG02475.1 KRG02476.1 GDHC01008458 GDHC01003474 JAQ10171.1 JAQ15155.1 CH940661 EDW71176.1 KRF85509.1 KRF85510.1 KRF85511.1 GEDC01025913 JAS11385.1 GEDC01001154 JAS36144.1 GECU01025895 JAS81811.1 ABLF02035238 CH940647 EDW70940.1 KRF85201.1 KRF85202.1 GDHF01028452 GDHF01018047 GDHF01002001 JAI23862.1 JAI34267.1 JAI50313.1 CP012525 ALC45046.1 LJIG01022819 KRT78556.1 GAMC01010411 JAB96144.1 GBHO01014271 GBRD01007904 JAG29333.1 JAG57917.1 GEDC01030302 GEDC01030086 GEDC01008426 GEDC01005369 GEDC01000164 JAS06996.1 JAS07212.1 JAS28872.1 JAS31929.1 JAS37134.1 PYGN01000259 PSN49875.1 GEDC01029100 GEDC01026518 GEDC01025608 GEDC01014560 GEDC01010316 GEDC01006295 GEDC01001363 JAS08198.1 JAS10780.1 JAS11690.1 JAS22738.1 JAS26982.1 JAS31003.1 JAS35935.1 GGMR01002706 MBY15325.1 CH916366 EDV97080.1 GGMS01017916 MBY87119.1 CH916368 EDW04294.1 MH172517 QAA95924.1 GGMS01010873 MBY80076.1 GAMC01017047 JAB89508.1 GECZ01011197 JAS58572.1 GBXI01015883 GBXI01015401 GBXI01002104 JAC98408.1 JAC98890.1 JAD12188.1 GAMC01010410 GAMC01010408 JAB96145.1 CH477809 EAT36018.1 KF828793 AIN44110.1 GAKP01001738 JAC57214.1 KY849640 ATY74521.1 GDHF01015383 GDHF01010110 GDHF01004300 JAI36931.1 JAI42204.1 JAI48014.1 GDQN01010299 JAT80755.1 DS231984 EDS30432.1 JRES01000502 KNC30573.1 GFDL01005899 JAV29146.1 ABLF02034227 ABLF02034231 ABLF02034236 GFDL01006011 JAV29034.1 GEBQ01011456 JAT28521.1 GFDL01006013 JAV29032.1 CP012523 ALC39759.1 GDHF01002644 JAI49670.1 GAKP01020912 GAKP01020911 JAC38040.1 KF828800 AIN44117.1 GBXI01010939 JAD03353.1 GGMR01013865 MBY26484.1 KY849643 ATY74524.1 GFXV01006246 MBW18051.1 JTDY01001309 KOB74240.1 GEDC01012756 JAS24542.1 ABLF02036668 ABLF02036673 GGMS01011330 MBY80533.1
KPI92893.1 NWSH01000663 PCG74941.1 ODYU01009275 SOQ53576.1 AGBW02011531 OWR46507.1 APGK01013174 KB738930 ENN82435.1 APGK01013159 APGK01013160 KB738929 ENN82439.1 KQ971338 KYB28165.1 KB632390 ERL94411.1 BT127729 AEE62691.1 KK854769 PTY18555.1 NEVH01021923 PNF19374.1 NEVH01006580 PNF37625.1 ACPB03010505 GBBI01003272 JAC15440.1 GDKW01002928 JAI53667.1 MF034805 AVT42171.1 GARF01000042 JAC88905.1 KK853032 KDR12258.1 KK853336 KDR08336.1 PYGN01000390 PSN47138.1 AY119083 AAM50943.1 AE014296 BT056236 AAF50035.2 ACL68683.1 AY075521 AAL68329.1 CM002912 KMY99032.1 CH480815 EDW41132.1 CH954178 EDV51440.1 CM000159 EDW94167.2 CH479240 EDW37581.1 GEDC01002932 JAS34366.1 KF828794 AIN44111.1 CH933809 EDW19319.1 CH902618 EDV39371.1 KPU78026.1 GEDC01011746 JAS25552.1 CH379069 EAL29640.1 OUUW01000004 SPP79199.1 OUUW01000002 SPP76781.1 CH963920 EDW77635.2 CH964062 EDW78857.2 SPP79198.1 CH479187 EDW39944.1 CH933807 EDW11002.1 KRG02475.1 KRG02476.1 GDHC01008458 GDHC01003474 JAQ10171.1 JAQ15155.1 CH940661 EDW71176.1 KRF85509.1 KRF85510.1 KRF85511.1 GEDC01025913 JAS11385.1 GEDC01001154 JAS36144.1 GECU01025895 JAS81811.1 ABLF02035238 CH940647 EDW70940.1 KRF85201.1 KRF85202.1 GDHF01028452 GDHF01018047 GDHF01002001 JAI23862.1 JAI34267.1 JAI50313.1 CP012525 ALC45046.1 LJIG01022819 KRT78556.1 GAMC01010411 JAB96144.1 GBHO01014271 GBRD01007904 JAG29333.1 JAG57917.1 GEDC01030302 GEDC01030086 GEDC01008426 GEDC01005369 GEDC01000164 JAS06996.1 JAS07212.1 JAS28872.1 JAS31929.1 JAS37134.1 PYGN01000259 PSN49875.1 GEDC01029100 GEDC01026518 GEDC01025608 GEDC01014560 GEDC01010316 GEDC01006295 GEDC01001363 JAS08198.1 JAS10780.1 JAS11690.1 JAS22738.1 JAS26982.1 JAS31003.1 JAS35935.1 GGMR01002706 MBY15325.1 CH916366 EDV97080.1 GGMS01017916 MBY87119.1 CH916368 EDW04294.1 MH172517 QAA95924.1 GGMS01010873 MBY80076.1 GAMC01017047 JAB89508.1 GECZ01011197 JAS58572.1 GBXI01015883 GBXI01015401 GBXI01002104 JAC98408.1 JAC98890.1 JAD12188.1 GAMC01010410 GAMC01010408 JAB96145.1 CH477809 EAT36018.1 KF828793 AIN44110.1 GAKP01001738 JAC57214.1 KY849640 ATY74521.1 GDHF01015383 GDHF01010110 GDHF01004300 JAI36931.1 JAI42204.1 JAI48014.1 GDQN01010299 JAT80755.1 DS231984 EDS30432.1 JRES01000502 KNC30573.1 GFDL01005899 JAV29146.1 ABLF02034227 ABLF02034231 ABLF02034236 GFDL01006011 JAV29034.1 GEBQ01011456 JAT28521.1 GFDL01006013 JAV29032.1 CP012523 ALC39759.1 GDHF01002644 JAI49670.1 GAKP01020912 GAKP01020911 JAC38040.1 KF828800 AIN44117.1 GBXI01010939 JAD03353.1 GGMR01013865 MBY26484.1 KY849643 ATY74524.1 GFXV01006246 MBW18051.1 JTDY01001309 KOB74240.1 GEDC01012756 JAS24542.1 ABLF02036668 ABLF02036673 GGMS01011330 MBY80533.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000019118
UP000007266
UP000030742
+ More
UP000235965 UP000015103 UP000027135 UP000245037 UP000000803 UP000001292 UP000008711 UP000002282 UP000192221 UP000008744 UP000009192 UP000007801 UP000001819 UP000268350 UP000079169 UP000007798 UP000008792 UP000007819 UP000192223 UP000092553 UP000001070 UP000008820 UP000002320 UP000037069 UP000037510
UP000235965 UP000015103 UP000027135 UP000245037 UP000000803 UP000001292 UP000008711 UP000002282 UP000192221 UP000008744 UP000009192 UP000007801 UP000001819 UP000268350 UP000079169 UP000007798 UP000008792 UP000007819 UP000192223 UP000092553 UP000001070 UP000008820 UP000002320 UP000037069 UP000037510
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A1D9C109
S4NIB2
A0A194PHU9
A0A194PIL7
A0A2A4JSF5
A0A2H1WKT2
+ More
A0A212EYH0 N6UL09 N6TPJ2 A0A139WK45 U4UVY2 J3JWW1 A0A2R7WJI2 A0A2J7PSR5 A0A2J7R9W6 T1I3H7 A0A023F2D5 A0A0P4VIF7 A0A2R4FXE8 A0A0U9HZ98 A0A067R385 A0A067QKC2 A0A2P8YSB2 Q8MS57 Q9VTL3 Q8SXX6 A0A0J9RTT3 B4HEW3 B3NGY3 B4PFL8 A0A1W4VU95 B4HAI3 A0A1B6E8X1 A0A158V6Y6 B4KUR3 B3M4A1 A0A1B6DIR7 Q2LZ69 A0A3B0JZT0 A0A3B0JVM2 A0A1S4EI61 B4N0M2 B4N3B8 A0A3B0K6S5 B4GQS2 B4KFN7 A0A146M787 B4MDB5 A0A1B6CDE3 A0A1B6EE46 A0A1B6I4E0 J9JXC1 B4LCP8 A0A1W4X256 A0A0K8V5S5 A0A0M4EG47 A0A0T6ATZ0 W8C2Y8 A0A0A9YE03 A0A1B6DT64 A0A2P8Z045 A0A1B6DAM8 A0A2S2NDX0 B4J2J5 A0A2S2RAU7 B4JD50 A0A3R5USX7 A0A2S2QQM8 W8AXI7 A0A1B6G7Z6 A0A0A1XNQ2 W8B4R5 Q16NT8 A0A158V4U5 A0A034WRH9 A0A2H4TES4 A0A0K8VTK0 A0A1E1W184 B0WLD9 A0A0L0CDT2 A0A1Q3FNT7 J9JSX9 A0A1Q3FNC5 A0A1B6LXW2 A0A1Q3FNE6 A0A0M3QTZ8 A0A0K8WFN7 A0A034V8F4 A0A158V1B5 A0A0A1WY89 A0A1W4UQ15 A0A2S2PAR8 A0A2H4TES0 A0A2H8TW86 A0A0L7LFT1 A0A1B6DFY5 J9K4G9 A0A2S2QS91
A0A212EYH0 N6UL09 N6TPJ2 A0A139WK45 U4UVY2 J3JWW1 A0A2R7WJI2 A0A2J7PSR5 A0A2J7R9W6 T1I3H7 A0A023F2D5 A0A0P4VIF7 A0A2R4FXE8 A0A0U9HZ98 A0A067R385 A0A067QKC2 A0A2P8YSB2 Q8MS57 Q9VTL3 Q8SXX6 A0A0J9RTT3 B4HEW3 B3NGY3 B4PFL8 A0A1W4VU95 B4HAI3 A0A1B6E8X1 A0A158V6Y6 B4KUR3 B3M4A1 A0A1B6DIR7 Q2LZ69 A0A3B0JZT0 A0A3B0JVM2 A0A1S4EI61 B4N0M2 B4N3B8 A0A3B0K6S5 B4GQS2 B4KFN7 A0A146M787 B4MDB5 A0A1B6CDE3 A0A1B6EE46 A0A1B6I4E0 J9JXC1 B4LCP8 A0A1W4X256 A0A0K8V5S5 A0A0M4EG47 A0A0T6ATZ0 W8C2Y8 A0A0A9YE03 A0A1B6DT64 A0A2P8Z045 A0A1B6DAM8 A0A2S2NDX0 B4J2J5 A0A2S2RAU7 B4JD50 A0A3R5USX7 A0A2S2QQM8 W8AXI7 A0A1B6G7Z6 A0A0A1XNQ2 W8B4R5 Q16NT8 A0A158V4U5 A0A034WRH9 A0A2H4TES4 A0A0K8VTK0 A0A1E1W184 B0WLD9 A0A0L0CDT2 A0A1Q3FNT7 J9JSX9 A0A1Q3FNC5 A0A1B6LXW2 A0A1Q3FNE6 A0A0M3QTZ8 A0A0K8WFN7 A0A034V8F4 A0A158V1B5 A0A0A1WY89 A0A1W4UQ15 A0A2S2PAR8 A0A2H4TES0 A0A2H8TW86 A0A0L7LFT1 A0A1B6DFY5 J9K4G9 A0A2S2QS91
PDB
6HCO
E-value=0.0294205,
Score=83
Ontologies
GO
Topology
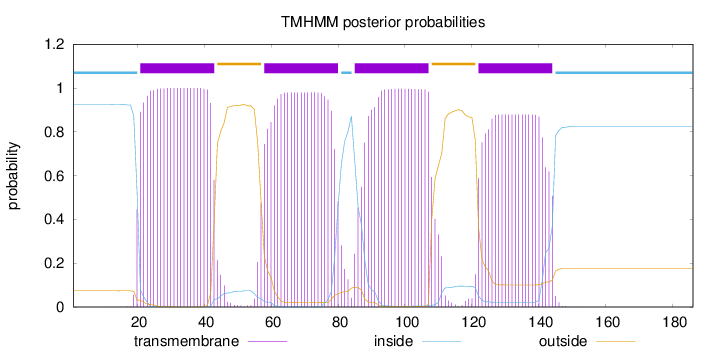
Length:
186
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.8385200000001
Exp number, first 60 AAs:
26.42062
Total prob of N-in:
0.92596
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 57
TMhelix
58 - 80
inside
81 - 84
TMhelix
85 - 107
outside
108 - 121
TMhelix
122 - 144
inside
145 - 186
Population Genetic Test Statistics
Pi
281.664122
Theta
186.261264
Tajima's D
1.287271
CLR
0.49859
CSRT
0.736113194340283
Interpretation
Uncertain
