Gene
KWMTBOMO07092
Pre Gene Modal
BGIBMGA010428
Annotation
PREDICTED:_zinc_finger_protein_177-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.317
Sequence
CDS
ATGAGGAATAACCTGTATAGACAGACCATGGACTCTATTTTGGAAGATTCTCCTTTAGATCTATCCACATCACTCCTTTCGAGAGTCCTTTTGCCAATATCTCCTCCGTCTGGACCAGAGGATGAACATAAATCGATATCGGGAGATTCCATGGCGTCGCTATCTGTTGGGTCATCGAATGAGCTGTTGGACATGACACCGAAAATCCAAAGAAGACCGACTAAGTCTGAGAGAGCGCTACTGCCTTGTGAAGTCTGCGGAAAGGCTTTTGATAGACCTTCTTTATTGAAACGACACATGAGGACACATACAGGAGAGAAGCCTCACGTGTGCATGGTGTGCAACAAAGGTTTCAGCACATCGAGCAGTCTGAACACACACAGAAGGATACACTCTGGCGAAAAGCCACATCAGTGTCAGCATTGCGGAAAACGATTCACGGCCAGCTCCAATCTATATTACCATCGAATGACCCACATAAAGTTAATTACTGAGTAA
Protein
MRNNLYRQTMDSILEDSPLDLSTSLLSRVLLPISPPSGPEDEHKSISGDSMASLSVGSSNELLDMTPKIQRRPTKSERALLPCEVCGKAFDRPSLLKRHMRTHTGEKPHVCMVCNKGFSTSSSLNTHRRIHSGEKPHQCQHCGKRFTASSNLYYHRMTHIKLITE
Summary
Uniprot
H9JLM7
A0A2H1WDX0
A0A2A4JTJ6
A0A212EMU8
A0A194PIM6
A0A3S2NU68
+ More
A0A194QN28 A0A336N012 A0A336LSN0 A0A1B0GJ59 A0A1J1IND2 W5JWI6 W5JCW1 A0A2M4CG16 A0A182Q3K1 A0A182GNW1 A0A182RGC4 Q7QG38 Q17NF2 A0A1S4EWM9 Q7QG37 A0A182Q7V8 A0A1B0GAN2 A0A1J1INC0 B0WBQ9 A0A182VA47 A0A182L5J6 A0A1S4JD60 A0A2J7PMS3 A0A067RTU1 A0A139WBT0 A0A0A9XY14 A0A1A9WEV1 A0A0T6B5J9 A0A1W4WPM1 U4UDQ2 N6TPY2 A0A1Y1KTJ1 A0A034V3N4 A0A0K8VYD1 A0A0A1XKK4 A0A182IYM6 W8BIK8 A0A2R7VWR4 A0A1B0C7T7 E0W2E1 A0A182L5J2 A0A182RGC7 A0A1A9ZX91 A0A1A9UIP8 A0A1B6EH12 A0A0R3P1I6 A0A2S2PNG4 A0A2S2QPN6 A0A2H8TUV0 A0A182J4X5 J9KAK7 A0A3B0KP31 A0A0R3NWJ2 A0A026VZV8 A0A087TJ71 A0A151XBV8 A0A195EC37 F4WNQ4 A0A195B8H5 A0A195FW15 T1JFB6 E1ZZE8 A0A151IPG2 A0A1E1X283 A0A2J7RJI9 B7PCU7 A0A2R5L806 A0A084VPK5
A0A194QN28 A0A336N012 A0A336LSN0 A0A1B0GJ59 A0A1J1IND2 W5JWI6 W5JCW1 A0A2M4CG16 A0A182Q3K1 A0A182GNW1 A0A182RGC4 Q7QG38 Q17NF2 A0A1S4EWM9 Q7QG37 A0A182Q7V8 A0A1B0GAN2 A0A1J1INC0 B0WBQ9 A0A182VA47 A0A182L5J6 A0A1S4JD60 A0A2J7PMS3 A0A067RTU1 A0A139WBT0 A0A0A9XY14 A0A1A9WEV1 A0A0T6B5J9 A0A1W4WPM1 U4UDQ2 N6TPY2 A0A1Y1KTJ1 A0A034V3N4 A0A0K8VYD1 A0A0A1XKK4 A0A182IYM6 W8BIK8 A0A2R7VWR4 A0A1B0C7T7 E0W2E1 A0A182L5J2 A0A182RGC7 A0A1A9ZX91 A0A1A9UIP8 A0A1B6EH12 A0A0R3P1I6 A0A2S2PNG4 A0A2S2QPN6 A0A2H8TUV0 A0A182J4X5 J9KAK7 A0A3B0KP31 A0A0R3NWJ2 A0A026VZV8 A0A087TJ71 A0A151XBV8 A0A195EC37 F4WNQ4 A0A195B8H5 A0A195FW15 T1JFB6 E1ZZE8 A0A151IPG2 A0A1E1X283 A0A2J7RJI9 B7PCU7 A0A2R5L806 A0A084VPK5
Pubmed
EMBL
BABH01005383
ODYU01007684
SOQ50684.1
NWSH01000640
PCG75096.1
AGBW02013784
+ More
OWR42825.1 KQ459603 KPI92903.1 RSAL01000079 RVE48693.1 KQ461194 KPJ06928.1 UFQS01003809 UFQT01003809 SSX15954.1 SSX35291.1 UFQT01000151 SSX20930.1 AJWK01019340 AJWK01019341 CVRI01000056 CRL01731.1 ADMH02000042 ETN68013.1 ADMH02001881 ETN60669.1 GGFL01000085 MBW64263.1 AXCN02000858 JXUM01077243 KQ562996 KXJ74683.1 AAAB01008839 EAA05906.4 CH477199 EAT48266.1 EAA05918.4 AXCN02000856 AXCN02000857 CCAG010011281 CRL01725.1 DS231880 EDS42594.1 NEVH01023960 PNF17619.1 KK852469 KDR23264.1 KQ971372 KYB25418.1 GBHO01021459 GDHC01007371 JAG22145.1 JAQ11258.1 LJIG01009661 KRT82633.1 KB632235 ERL90428.1 APGK01058983 APGK01058984 KB741292 ENN70366.1 GEZM01075889 JAV63898.1 GAKP01022220 JAC36732.1 GDHF01008470 JAI43844.1 GBXI01002810 JAD11482.1 GAMC01009702 JAB96853.1 KK854103 PTY11341.1 JXJN01031103 DS235877 EEB19797.1 GEDC01000134 JAS37164.1 CH475516 KRT05365.1 GGMR01018326 MBY30945.1 GGMS01010543 MBY79746.1 GFXV01006220 MBW18025.1 ABLF02033911 OUUW01000010 SPP85598.1 KRT05364.1 KK107558 EZA48991.1 KK115458 KFM65160.1 KQ982316 KYQ57845.1 KQ979074 KYN22788.1 GL888237 EGI64247.1 KQ976565 KYM80542.1 KQ981215 KYN44512.1 JH432147 GL435343 EFN73440.1 KQ976892 KYN07228.1 GFAC01005808 JAT93380.1 NEVH01002992 PNF40997.1 ABJB010350451 ABJB010596856 ABJB010803269 DS686235 EEC04419.1 GGLE01001483 MBY05609.1 ATLV01015011 KE524999 KFB39899.1
OWR42825.1 KQ459603 KPI92903.1 RSAL01000079 RVE48693.1 KQ461194 KPJ06928.1 UFQS01003809 UFQT01003809 SSX15954.1 SSX35291.1 UFQT01000151 SSX20930.1 AJWK01019340 AJWK01019341 CVRI01000056 CRL01731.1 ADMH02000042 ETN68013.1 ADMH02001881 ETN60669.1 GGFL01000085 MBW64263.1 AXCN02000858 JXUM01077243 KQ562996 KXJ74683.1 AAAB01008839 EAA05906.4 CH477199 EAT48266.1 EAA05918.4 AXCN02000856 AXCN02000857 CCAG010011281 CRL01725.1 DS231880 EDS42594.1 NEVH01023960 PNF17619.1 KK852469 KDR23264.1 KQ971372 KYB25418.1 GBHO01021459 GDHC01007371 JAG22145.1 JAQ11258.1 LJIG01009661 KRT82633.1 KB632235 ERL90428.1 APGK01058983 APGK01058984 KB741292 ENN70366.1 GEZM01075889 JAV63898.1 GAKP01022220 JAC36732.1 GDHF01008470 JAI43844.1 GBXI01002810 JAD11482.1 GAMC01009702 JAB96853.1 KK854103 PTY11341.1 JXJN01031103 DS235877 EEB19797.1 GEDC01000134 JAS37164.1 CH475516 KRT05365.1 GGMR01018326 MBY30945.1 GGMS01010543 MBY79746.1 GFXV01006220 MBW18025.1 ABLF02033911 OUUW01000010 SPP85598.1 KRT05364.1 KK107558 EZA48991.1 KK115458 KFM65160.1 KQ982316 KYQ57845.1 KQ979074 KYN22788.1 GL888237 EGI64247.1 KQ976565 KYM80542.1 KQ981215 KYN44512.1 JH432147 GL435343 EFN73440.1 KQ976892 KYN07228.1 GFAC01005808 JAT93380.1 NEVH01002992 PNF40997.1 ABJB010350451 ABJB010596856 ABJB010803269 DS686235 EEC04419.1 GGLE01001483 MBY05609.1 ATLV01015011 KE524999 KFB39899.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
+ More
UP000092461 UP000183832 UP000000673 UP000075886 UP000069940 UP000249989 UP000075900 UP000007062 UP000008820 UP000092444 UP000002320 UP000075903 UP000075882 UP000235965 UP000027135 UP000007266 UP000091820 UP000192223 UP000030742 UP000019118 UP000075880 UP000092460 UP000009046 UP000092445 UP000078200 UP000001819 UP000007819 UP000268350 UP000053097 UP000054359 UP000075809 UP000078492 UP000007755 UP000078540 UP000078541 UP000000311 UP000078542 UP000001555 UP000030765
UP000092461 UP000183832 UP000000673 UP000075886 UP000069940 UP000249989 UP000075900 UP000007062 UP000008820 UP000092444 UP000002320 UP000075903 UP000075882 UP000235965 UP000027135 UP000007266 UP000091820 UP000192223 UP000030742 UP000019118 UP000075880 UP000092460 UP000009046 UP000092445 UP000078200 UP000001819 UP000007819 UP000268350 UP000053097 UP000054359 UP000075809 UP000078492 UP000007755 UP000078540 UP000078541 UP000000311 UP000078542 UP000001555 UP000030765
PRIDE
Pfam
PF00096 zf-C2H2
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JLM7
A0A2H1WDX0
A0A2A4JTJ6
A0A212EMU8
A0A194PIM6
A0A3S2NU68
+ More
A0A194QN28 A0A336N012 A0A336LSN0 A0A1B0GJ59 A0A1J1IND2 W5JWI6 W5JCW1 A0A2M4CG16 A0A182Q3K1 A0A182GNW1 A0A182RGC4 Q7QG38 Q17NF2 A0A1S4EWM9 Q7QG37 A0A182Q7V8 A0A1B0GAN2 A0A1J1INC0 B0WBQ9 A0A182VA47 A0A182L5J6 A0A1S4JD60 A0A2J7PMS3 A0A067RTU1 A0A139WBT0 A0A0A9XY14 A0A1A9WEV1 A0A0T6B5J9 A0A1W4WPM1 U4UDQ2 N6TPY2 A0A1Y1KTJ1 A0A034V3N4 A0A0K8VYD1 A0A0A1XKK4 A0A182IYM6 W8BIK8 A0A2R7VWR4 A0A1B0C7T7 E0W2E1 A0A182L5J2 A0A182RGC7 A0A1A9ZX91 A0A1A9UIP8 A0A1B6EH12 A0A0R3P1I6 A0A2S2PNG4 A0A2S2QPN6 A0A2H8TUV0 A0A182J4X5 J9KAK7 A0A3B0KP31 A0A0R3NWJ2 A0A026VZV8 A0A087TJ71 A0A151XBV8 A0A195EC37 F4WNQ4 A0A195B8H5 A0A195FW15 T1JFB6 E1ZZE8 A0A151IPG2 A0A1E1X283 A0A2J7RJI9 B7PCU7 A0A2R5L806 A0A084VPK5
A0A194QN28 A0A336N012 A0A336LSN0 A0A1B0GJ59 A0A1J1IND2 W5JWI6 W5JCW1 A0A2M4CG16 A0A182Q3K1 A0A182GNW1 A0A182RGC4 Q7QG38 Q17NF2 A0A1S4EWM9 Q7QG37 A0A182Q7V8 A0A1B0GAN2 A0A1J1INC0 B0WBQ9 A0A182VA47 A0A182L5J6 A0A1S4JD60 A0A2J7PMS3 A0A067RTU1 A0A139WBT0 A0A0A9XY14 A0A1A9WEV1 A0A0T6B5J9 A0A1W4WPM1 U4UDQ2 N6TPY2 A0A1Y1KTJ1 A0A034V3N4 A0A0K8VYD1 A0A0A1XKK4 A0A182IYM6 W8BIK8 A0A2R7VWR4 A0A1B0C7T7 E0W2E1 A0A182L5J2 A0A182RGC7 A0A1A9ZX91 A0A1A9UIP8 A0A1B6EH12 A0A0R3P1I6 A0A2S2PNG4 A0A2S2QPN6 A0A2H8TUV0 A0A182J4X5 J9KAK7 A0A3B0KP31 A0A0R3NWJ2 A0A026VZV8 A0A087TJ71 A0A151XBV8 A0A195EC37 F4WNQ4 A0A195B8H5 A0A195FW15 T1JFB6 E1ZZE8 A0A151IPG2 A0A1E1X283 A0A2J7RJI9 B7PCU7 A0A2R5L806 A0A084VPK5
PDB
5V3G
E-value=6.46601e-16,
Score=200
Ontologies
GO
PANTHER
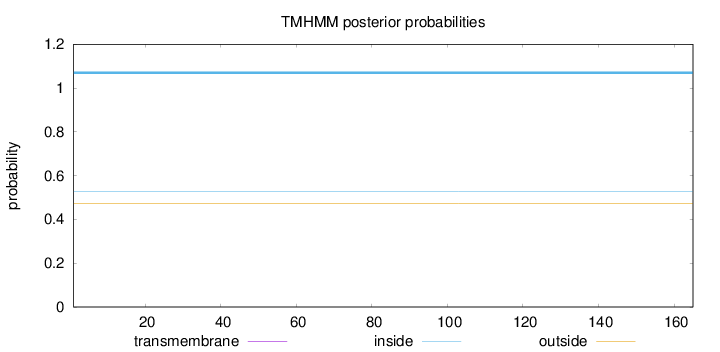
Topology
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.52641
inside
1 - 165
Population Genetic Test Statistics
Pi
237.058213
Theta
158.295206
Tajima's D
0.865495
CLR
112.567255
CSRT
0.624768761561922
Interpretation
Uncertain
