Gene
KWMTBOMO07091
Annotation
zinc_finger_protein_[Danaus_plexippus]
Transcription factor
Location in the cell
Mitochondrial Reliability : 1.525 Nuclear Reliability : 2.087
Sequence
CDS
ATGAGCACAAAAGAGAAGCCTCATAAGTGCACGCTCTGCTCCAAGTCGTTCCCGACCCCGGGGGACCTCAAGTCGCACATGTACGTCCACAGCGGATCCTGGCCCTACAAATGTCACATTTGTTCTAGAGGATTCTCGAAGCACACTAACTTGAAGAACCATTTGTTCCTTCATACCGCGAAACATCAAAGGAACAGTGCCAGGGACGCAACTTGA
Protein
MSTKEKPHKCTLCSKSFPTPGDLKSHMYVHSGSWPYKCHICSRGFSKHTNLKNHLFLHTAKHQRNSARDAT
Summary
Uniprot
A0A2H1VVE3
A0A2A4JTJ6
A0A194PIM6
A0A3S2NU68
A0A212EMU8
A0A182VUW3
+ More
T1H1P0 A0A1V9Y3A4 A0A194QN28 A0A336LSN0 A0A0N8E7I3 A0A162S347 A0A1J1IPS5 A0A0P5MU04 A0A0P5LGY9 B7PCU7 A0A2P8XF91 A0A0P5KQT4 N6TPY2 A0A1B0GJ59 A0A0P5HVT0 A0A0N8A4E5 A0A0P5ZIC1 A0A0P5PXW1 A0A0P5Q055 A0A0P5WDK5 A0A0P5KHE8 A0A0T6B5J9 A0A084VPK0 A0A0P5K5U8 A0A0P5KHW3 A0A139WBT0 A0A0P5EFB3 A0A0P5IZN4 A0A0N7ZN58 A0A0P6F704 A0A0P6E554 A0A0P5UD16 A0A0P4ZIA2 A0A0P5SRQ4 A0A0N8E3H2 A0A0P5KXB6 E9G187 A0A0P5L431 V5H995 A0A0P5IGT3 A0A2R5L806 A0A1B6EH12 A0A1E1X283 A0A0P5CKJ4 A0A0P5G4G1 A0A0P6I2X5 A0A0A9XY14 A0A182YMG0 A0A0P6BD76 U4UDQ2 B0WBQ9 A0A067RTU1 E0W2E1 T1HG44 A0A2H8TUV0 A0A1B6IAX4 A0A0P5MTK0 A0A084VPK6 A0A2S2QPN6 Q7QG37 J9KAK7 A0A1B6GXQ6 A0A1B6EXD1 W5JWI6 A0A2S2PNG4 A0A1Y1KTJ1 A0A1A9Y1Y6 A0A182MVZ2 A0A182IE74 A0A1W4WPM1 A0A182TXM6 A0A0P5JSX8 A0A182GNW1 A0A182V0K8 A0A182IYM6 A0A1S3DE05 A0A1A9ZX91 A0A1A9UIP8 A0A182L5J2 T1JFB6 A0A087TJ71 A0A182Q7V8 A0A182RGC4 A0A1B0C7T7 A0A1S4EWM9 Q17NF2 A0A1J1INC0 A0A1J1IND2 A0A182RGC7 A0A1S4JD60 A0A182UBI6 A0A182J4X5 Q7QG38 A0A1A9WEV1
T1H1P0 A0A1V9Y3A4 A0A194QN28 A0A336LSN0 A0A0N8E7I3 A0A162S347 A0A1J1IPS5 A0A0P5MU04 A0A0P5LGY9 B7PCU7 A0A2P8XF91 A0A0P5KQT4 N6TPY2 A0A1B0GJ59 A0A0P5HVT0 A0A0N8A4E5 A0A0P5ZIC1 A0A0P5PXW1 A0A0P5Q055 A0A0P5WDK5 A0A0P5KHE8 A0A0T6B5J9 A0A084VPK0 A0A0P5K5U8 A0A0P5KHW3 A0A139WBT0 A0A0P5EFB3 A0A0P5IZN4 A0A0N7ZN58 A0A0P6F704 A0A0P6E554 A0A0P5UD16 A0A0P4ZIA2 A0A0P5SRQ4 A0A0N8E3H2 A0A0P5KXB6 E9G187 A0A0P5L431 V5H995 A0A0P5IGT3 A0A2R5L806 A0A1B6EH12 A0A1E1X283 A0A0P5CKJ4 A0A0P5G4G1 A0A0P6I2X5 A0A0A9XY14 A0A182YMG0 A0A0P6BD76 U4UDQ2 B0WBQ9 A0A067RTU1 E0W2E1 T1HG44 A0A2H8TUV0 A0A1B6IAX4 A0A0P5MTK0 A0A084VPK6 A0A2S2QPN6 Q7QG37 J9KAK7 A0A1B6GXQ6 A0A1B6EXD1 W5JWI6 A0A2S2PNG4 A0A1Y1KTJ1 A0A1A9Y1Y6 A0A182MVZ2 A0A182IE74 A0A1W4WPM1 A0A182TXM6 A0A0P5JSX8 A0A182GNW1 A0A182V0K8 A0A182IYM6 A0A1S3DE05 A0A1A9ZX91 A0A1A9UIP8 A0A182L5J2 T1JFB6 A0A087TJ71 A0A182Q7V8 A0A182RGC4 A0A1B0C7T7 A0A1S4EWM9 Q17NF2 A0A1J1INC0 A0A1J1IND2 A0A182RGC7 A0A1S4JD60 A0A182UBI6 A0A182J4X5 Q7QG38 A0A1A9WEV1
Pubmed
EMBL
ODYU01004660
SOQ44783.1
NWSH01000640
PCG75096.1
KQ459603
KPI92903.1
+ More
RSAL01000079 RVE48693.1 AGBW02013784 OWR42825.1 CAQQ02157671 CAQQ02157672 MNPL01000182 OQR80247.1 KQ461194 KPJ06928.1 UFQT01000151 SSX20930.1 GDIQ01054204 JAN40533.1 LRGB01000084 KZS20978.1 CVRI01000056 CRL01722.1 GDIQ01173895 JAK77830.1 GDIQ01173894 JAK77831.1 ABJB010350451 ABJB010596856 ABJB010803269 DS686235 EEC04419.1 PYGN01002357 PSN30669.1 GDIQ01181999 JAK69726.1 APGK01058983 APGK01058984 KB741292 ENN70366.1 AJWK01019340 AJWK01019341 GDIQ01222289 JAK29436.1 GDIP01183577 JAJ39825.1 GDIP01046645 JAM57070.1 GDIQ01124074 JAL27652.1 GDIQ01137312 GDIQ01121622 GDIQ01113835 GDIP01130692 JAL30104.1 JAL73022.1 GDIP01088339 JAM15376.1 GDIQ01209689 JAK42036.1 LJIG01009661 KRT82633.1 ATLV01015007 KE524999 KFB39894.1 GDIQ01188745 JAK62980.1 GDIQ01183854 JAK67871.1 KQ971372 KYB25418.1 GDIP01149130 JAJ74272.1 GDIQ01208292 JAK43433.1 GDIP01229073 JAI94328.1 GDIQ01052407 JAN42330.1 GDIQ01082280 JAN12457.1 GDIP01132041 JAL71673.1 GDIP01212434 JAJ10968.1 GDIP01136268 JAL67446.1 GDIQ01065492 JAN29245.1 GDIQ01177852 JAK73873.1 GL732529 EFX86682.1 GDIQ01180705 JAK71020.1 GANP01010724 JAB73744.1 GDIQ01217145 JAK34580.1 GGLE01001483 MBY05609.1 GEDC01000134 JAS37164.1 GFAC01005808 JAT93380.1 GDIP01184928 JAJ38474.1 GDIQ01246422 JAK05303.1 GDIQ01011310 JAN83427.1 GBHO01021459 GDHC01007371 JAG22145.1 JAQ11258.1 GDIP01016568 JAM87147.1 KB632235 ERL90428.1 DS231880 EDS42594.1 KK852469 KDR23264.1 DS235877 EEB19797.1 ACPB03001144 GFXV01006220 MBW18025.1 GECU01023696 JAS84010.1 GDIQ01152064 JAK99661.1 ATLV01015011 KFB39900.1 GGMS01010543 MBY79746.1 AAAB01008839 EAA05918.4 ABLF02033911 GECZ01002554 JAS67215.1 GECZ01027144 JAS42625.1 ADMH02000042 ETN68013.1 GGMR01018326 MBY30945.1 GEZM01075889 JAV63898.1 AXCM01002127 APCN01004575 GDIQ01194446 JAK57279.1 JXUM01077243 KQ562996 KXJ74683.1 JH432147 KK115458 KFM65160.1 AXCN02000856 AXCN02000857 JXJN01031103 CH477199 EAT48266.1 CRL01725.1 CRL01731.1 EAA05906.4
RSAL01000079 RVE48693.1 AGBW02013784 OWR42825.1 CAQQ02157671 CAQQ02157672 MNPL01000182 OQR80247.1 KQ461194 KPJ06928.1 UFQT01000151 SSX20930.1 GDIQ01054204 JAN40533.1 LRGB01000084 KZS20978.1 CVRI01000056 CRL01722.1 GDIQ01173895 JAK77830.1 GDIQ01173894 JAK77831.1 ABJB010350451 ABJB010596856 ABJB010803269 DS686235 EEC04419.1 PYGN01002357 PSN30669.1 GDIQ01181999 JAK69726.1 APGK01058983 APGK01058984 KB741292 ENN70366.1 AJWK01019340 AJWK01019341 GDIQ01222289 JAK29436.1 GDIP01183577 JAJ39825.1 GDIP01046645 JAM57070.1 GDIQ01124074 JAL27652.1 GDIQ01137312 GDIQ01121622 GDIQ01113835 GDIP01130692 JAL30104.1 JAL73022.1 GDIP01088339 JAM15376.1 GDIQ01209689 JAK42036.1 LJIG01009661 KRT82633.1 ATLV01015007 KE524999 KFB39894.1 GDIQ01188745 JAK62980.1 GDIQ01183854 JAK67871.1 KQ971372 KYB25418.1 GDIP01149130 JAJ74272.1 GDIQ01208292 JAK43433.1 GDIP01229073 JAI94328.1 GDIQ01052407 JAN42330.1 GDIQ01082280 JAN12457.1 GDIP01132041 JAL71673.1 GDIP01212434 JAJ10968.1 GDIP01136268 JAL67446.1 GDIQ01065492 JAN29245.1 GDIQ01177852 JAK73873.1 GL732529 EFX86682.1 GDIQ01180705 JAK71020.1 GANP01010724 JAB73744.1 GDIQ01217145 JAK34580.1 GGLE01001483 MBY05609.1 GEDC01000134 JAS37164.1 GFAC01005808 JAT93380.1 GDIP01184928 JAJ38474.1 GDIQ01246422 JAK05303.1 GDIQ01011310 JAN83427.1 GBHO01021459 GDHC01007371 JAG22145.1 JAQ11258.1 GDIP01016568 JAM87147.1 KB632235 ERL90428.1 DS231880 EDS42594.1 KK852469 KDR23264.1 DS235877 EEB19797.1 ACPB03001144 GFXV01006220 MBW18025.1 GECU01023696 JAS84010.1 GDIQ01152064 JAK99661.1 ATLV01015011 KFB39900.1 GGMS01010543 MBY79746.1 AAAB01008839 EAA05918.4 ABLF02033911 GECZ01002554 JAS67215.1 GECZ01027144 JAS42625.1 ADMH02000042 ETN68013.1 GGMR01018326 MBY30945.1 GEZM01075889 JAV63898.1 AXCM01002127 APCN01004575 GDIQ01194446 JAK57279.1 JXUM01077243 KQ562996 KXJ74683.1 JH432147 KK115458 KFM65160.1 AXCN02000856 AXCN02000857 JXJN01031103 CH477199 EAT48266.1 CRL01725.1 CRL01731.1 EAA05906.4
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000075920
UP000015102
+ More
UP000192247 UP000053240 UP000076858 UP000183832 UP000001555 UP000245037 UP000019118 UP000092461 UP000030765 UP000007266 UP000000305 UP000076408 UP000030742 UP000002320 UP000027135 UP000009046 UP000015103 UP000007062 UP000007819 UP000000673 UP000092443 UP000075883 UP000075840 UP000192223 UP000075902 UP000069940 UP000249989 UP000075903 UP000075880 UP000079169 UP000092445 UP000078200 UP000075882 UP000054359 UP000075886 UP000075900 UP000092460 UP000008820 UP000091820
UP000192247 UP000053240 UP000076858 UP000183832 UP000001555 UP000245037 UP000019118 UP000092461 UP000030765 UP000007266 UP000000305 UP000076408 UP000030742 UP000002320 UP000027135 UP000009046 UP000015103 UP000007062 UP000007819 UP000000673 UP000092443 UP000075883 UP000075840 UP000192223 UP000075902 UP000069940 UP000249989 UP000075903 UP000075880 UP000079169 UP000092445 UP000078200 UP000075882 UP000054359 UP000075886 UP000075900 UP000092460 UP000008820 UP000091820
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
A0A2H1VVE3
A0A2A4JTJ6
A0A194PIM6
A0A3S2NU68
A0A212EMU8
A0A182VUW3
+ More
T1H1P0 A0A1V9Y3A4 A0A194QN28 A0A336LSN0 A0A0N8E7I3 A0A162S347 A0A1J1IPS5 A0A0P5MU04 A0A0P5LGY9 B7PCU7 A0A2P8XF91 A0A0P5KQT4 N6TPY2 A0A1B0GJ59 A0A0P5HVT0 A0A0N8A4E5 A0A0P5ZIC1 A0A0P5PXW1 A0A0P5Q055 A0A0P5WDK5 A0A0P5KHE8 A0A0T6B5J9 A0A084VPK0 A0A0P5K5U8 A0A0P5KHW3 A0A139WBT0 A0A0P5EFB3 A0A0P5IZN4 A0A0N7ZN58 A0A0P6F704 A0A0P6E554 A0A0P5UD16 A0A0P4ZIA2 A0A0P5SRQ4 A0A0N8E3H2 A0A0P5KXB6 E9G187 A0A0P5L431 V5H995 A0A0P5IGT3 A0A2R5L806 A0A1B6EH12 A0A1E1X283 A0A0P5CKJ4 A0A0P5G4G1 A0A0P6I2X5 A0A0A9XY14 A0A182YMG0 A0A0P6BD76 U4UDQ2 B0WBQ9 A0A067RTU1 E0W2E1 T1HG44 A0A2H8TUV0 A0A1B6IAX4 A0A0P5MTK0 A0A084VPK6 A0A2S2QPN6 Q7QG37 J9KAK7 A0A1B6GXQ6 A0A1B6EXD1 W5JWI6 A0A2S2PNG4 A0A1Y1KTJ1 A0A1A9Y1Y6 A0A182MVZ2 A0A182IE74 A0A1W4WPM1 A0A182TXM6 A0A0P5JSX8 A0A182GNW1 A0A182V0K8 A0A182IYM6 A0A1S3DE05 A0A1A9ZX91 A0A1A9UIP8 A0A182L5J2 T1JFB6 A0A087TJ71 A0A182Q7V8 A0A182RGC4 A0A1B0C7T7 A0A1S4EWM9 Q17NF2 A0A1J1INC0 A0A1J1IND2 A0A182RGC7 A0A1S4JD60 A0A182UBI6 A0A182J4X5 Q7QG38 A0A1A9WEV1
T1H1P0 A0A1V9Y3A4 A0A194QN28 A0A336LSN0 A0A0N8E7I3 A0A162S347 A0A1J1IPS5 A0A0P5MU04 A0A0P5LGY9 B7PCU7 A0A2P8XF91 A0A0P5KQT4 N6TPY2 A0A1B0GJ59 A0A0P5HVT0 A0A0N8A4E5 A0A0P5ZIC1 A0A0P5PXW1 A0A0P5Q055 A0A0P5WDK5 A0A0P5KHE8 A0A0T6B5J9 A0A084VPK0 A0A0P5K5U8 A0A0P5KHW3 A0A139WBT0 A0A0P5EFB3 A0A0P5IZN4 A0A0N7ZN58 A0A0P6F704 A0A0P6E554 A0A0P5UD16 A0A0P4ZIA2 A0A0P5SRQ4 A0A0N8E3H2 A0A0P5KXB6 E9G187 A0A0P5L431 V5H995 A0A0P5IGT3 A0A2R5L806 A0A1B6EH12 A0A1E1X283 A0A0P5CKJ4 A0A0P5G4G1 A0A0P6I2X5 A0A0A9XY14 A0A182YMG0 A0A0P6BD76 U4UDQ2 B0WBQ9 A0A067RTU1 E0W2E1 T1HG44 A0A2H8TUV0 A0A1B6IAX4 A0A0P5MTK0 A0A084VPK6 A0A2S2QPN6 Q7QG37 J9KAK7 A0A1B6GXQ6 A0A1B6EXD1 W5JWI6 A0A2S2PNG4 A0A1Y1KTJ1 A0A1A9Y1Y6 A0A182MVZ2 A0A182IE74 A0A1W4WPM1 A0A182TXM6 A0A0P5JSX8 A0A182GNW1 A0A182V0K8 A0A182IYM6 A0A1S3DE05 A0A1A9ZX91 A0A1A9UIP8 A0A182L5J2 T1JFB6 A0A087TJ71 A0A182Q7V8 A0A182RGC4 A0A1B0C7T7 A0A1S4EWM9 Q17NF2 A0A1J1INC0 A0A1J1IND2 A0A182RGC7 A0A1S4JD60 A0A182UBI6 A0A182J4X5 Q7QG38 A0A1A9WEV1
PDB
2LCE
E-value=1.05021e-06,
Score=119
Ontologies
GO
PANTHER
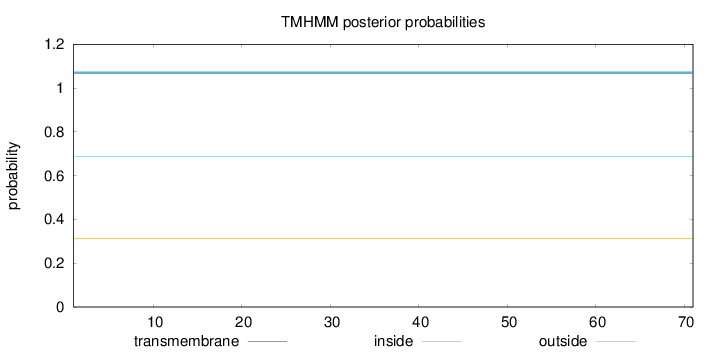
Topology
Length:
71
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0004
Exp number, first 60 AAs:
0.0004
Total prob of N-in:
0.68620
inside
1 - 71
Population Genetic Test Statistics
Pi
177.573382
Theta
164.962522
Tajima's D
0.140578
CLR
1.890485
CSRT
0.405929703514824
Interpretation
Uncertain
