Gene
KWMTBOMO07088
Pre Gene Modal
BGIBMGA010528
Annotation
PREDICTED:_sodium-dependent_nutrient_amino_acid_transporter_1-like_isoform_X1_[Amyelois_transitella]
Full name
Transporter
Location in the cell
PlasmaMembrane Reliability : 4.834
Sequence
CDS
ATGGCAGTTGTGACACGACCGTGCGGTGACTTCCGTTGGATACGTAACAGCGCCTGCAGGAAGGGCACATCGCGAAATGGCTGGAATGTAAGATCAGCGTGTGACTTCGTCAAGACACAAATATGTACTGTGGCCCTTTCACTGACGCTCTTCAACTGCGCGCGTCTACCGAAAGAGGCATTCAAGTATGGCGCTGTTCCGTACCTGGTGGTCTACACATTCTGGCTTCTGGCCGTTGGGCTGCCAACGACTTTGCTGGAGCTCGCCATGGGACAGCTTAGTCAGCAAGATCCGGTGGGAGTTTGGAGAGCTGTTCCTATATTAAGAGGTGTGGGTCATCTAAAAGTTTTGACTTCGTATATATGCTGTGTTTATTACATGATGTACGTAGCTTTGGCCCTGGCCTTCCTCGTTTGGATAGGGAAAGGCACTTTTCCGCTGAGAGACTGTACTAGGCTTCAAATGACGCCGAACGGATACGAAAACAAAATGAACGCATCTGAATGTTTCAAATCAACATTCTTGTCACCTTTCACGGACTATCCACAATATTTGGGAATCATGGCGATACTTATCTTCATTTTATGGTTTTTCTTGCCTATACTATTATATCGTCTTCGAAAAACGCTTAAAGTAGCTATGACTGTGTTAGTGCCATCTGTCGTACTATTTGCTGCATTGCTAGCTACGTTTCTGGCCAAGAGTGACGTGTTGAATGCCATGTTCGAATCTTGCGAGCAATGGACGCCGTTATCAGAGCCGTACATTTGGCACAGCGCTTTAGTGCAGGCGTTGCTGTCCACGCACATATCTGGAGGCTACCTGATCAGTTCCGGGGGAACGATTTATAGGCATTCGGATGTGCGATGGACGTCAGCATGCGTTATCATGACCAGTATATTCTCGGGTTGGCTTTGGGTGCTTCTCTGGGAGTCTATCGACGCAGAAAGCAAGAAGAACACTGATTTCGTATCAATCCTCGTGTTAGTTTATCAGTCTTCTATATCAGAGAGGAGATCGAAGCAGTGGCCTTTGCTGGCTTATGGAGTTATATTCTTATCTGGTATCACCACTATGGTTACCCTCCTGTTTCCTGTCTTCGACAAACTACATCGTCTCACCGGTGACCACTGGAGGTTGTTCGCCAGCGCGTCTTCAGCTGTTGGTACAGCCTTCACCGTGGCAGTGTTGGCCCGAGGTCTTGAAGTCGCCACGATGCTGGACGATTTGGTCATTCCGCTGCTAACAACATTCACCACGGCCGTTGAAGTGGTCGGATTTGTTTTTATTTATGGTTGGTGCTACCTCACGATTGACATAGAGTTTCTCACTGGCATGAAACTACCCTGTTTCTGGATAGCGACGTGGTGGTGTACGCCTATATTAGTGCTGGGTACCACTGGCTGGTGGCTTCGAGCTCTGCTCCGAGCTACTTGGGGCCAGGAGGAGACGCTGTGGCCTCTAGTCGGAGCTTTCGCGGGGATTCTCATAGTCATGGTCGTCATCGCCGCTGTGGCCGTGGCTAAAGAAGAACAATTCAATTTAACTTCGAAAATATCATCGGCCTTCAAGTCGTCGCGGCTGTGGGGTCCCGAGGAGCCGATGGCGCGGTACGTTTGGATGTCGCAGCGTTACCTCAACGACACGCTCAGCAGCACCAATGACGTCAGCTACGAACGGCCCGACAGCGACACCGCCCACAACAAATACAAAGAAAACTGGCTTAGCCATAAAGATAAATACCAAAAAGAATACGATTATTACTCGAGGATCGTCGCGACGAAAACTCCAAGCAAAGCCTCCAAAGAGACATACCCAGCGCACACTATCCCAAGAAGCGAAACGAAGGTTCCAGAAGAGAAGTTCCGGTCGCCCAACATATGCATCGCTGAGGCCCAAATCGGGGGCCCCACGAATTGCAACTGCAACAGACATTTCAAACTAAACGTACCTAAAGATCTAAGGAACAATGAAGTTACGACGAGCTTATGA
Protein
MAVVTRPCGDFRWIRNSACRKGTSRNGWNVRSACDFVKTQICTVALSLTLFNCARLPKEAFKYGAVPYLVVYTFWLLAVGLPTTLLELAMGQLSQQDPVGVWRAVPILRGVGHLKVLTSYICCVYYMMYVALALAFLVWIGKGTFPLRDCTRLQMTPNGYENKMNASECFKSTFLSPFTDYPQYLGIMAILIFILWFFLPILLYRLRKTLKVAMTVLVPSVVLFAALLATFLAKSDVLNAMFESCEQWTPLSEPYIWHSALVQALLSTHISGGYLISSGGTIYRHSDVRWTSACVIMTSIFSGWLWVLLWESIDAESKKNTDFVSILVLVYQSSISERRSKQWPLLAYGVIFLSGITTMVTLLFPVFDKLHRLTGDHWRLFASASSAVGTAFTVAVLARGLEVATMLDDLVIPLLTTFTTAVEVVGFVFIYGWCYLTIDIEFLTGMKLPCFWIATWWCTPILVLGTTGWWLRALLRATWGQEETLWPLVGAFAGILIVMVVIAAVAVAKEEQFNLTSKISSAFKSSRLWGPEEPMARYVWMSQRYLNDTLSSTNDVSYERPDSDTAHNKYKENWLSHKDKYQKEYDYYSRIVATKTPSKASKETYPAHTIPRSETKVPEEKFRSPNICIAEAQIGGPTNCNCNRHFKLNVPKDLRNNEVTTSL
Summary
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Uniprot
H9JLX6
A0A2A4JQ39
A0A2H1VKR8
A0A194PIN1
A0A194QNJ1
A0A2H1WUH2
+ More
A0A212EQK9 A0A194PFI0 A0A194R384 A0A2W1BHG8 A0A2A4JUW4 A0A2A4JU52 A0A069DWH7 A0A146LQK9 A0A146MEM0 A0A2H8TMM4 T1HU77 A0A2S2PQU7 A0A2S2PMX6 A0A1B6DBA5 J9KES3 A0A1B6CRS2 A0A2S2QZ54 X2D4J3 A0A2J7Q3G4 D6WED3 B6CY69 A0A1B6LCC7 A0A1B6F5R4 A0A1Y1MI14 A0A1Y1MIE3 A0A1Y1ML21 A0A1B6IXX6 A0A1B6GDB9 N6U4N6 A0A336MYS6 A0A1L8DJV8 A0A1L8DJV5 A0A034VAZ8 A0A336L0X2 A0A0L0CIV0 A0A0R1E314 B4PJW1 A0A1I8PP83 B4MMQ9 B4KZ22 A0A1I8MQX4 B4J247 A0A1B0EY28 A0A0Q5UJS4 Q8IQQ6 C7LAC7 B3NHW0 A0A0Q9XPD1 Q9VVG3 A0A0J9UN26 B5DQ44 A0A3B0KG05 Q9U975 Q8IQQ7 A0A0M3QWZ4 A0A0P8YGX5 A0A0J9RYC3 A0A1I8PP26 C7LA83 A0A336LX44 B4QNU2 B4HKR7 B3M6C3 A0A1A9VYS9 A0A1W4W4W9 A0A1B0C4T3 A0A336KAS2 A0A1W4W4B2 A0A1B0FAG9 A0A1A9WEB0 A0A0K8TQT5 A0A182N207 B4H6L6 A0A084WCY0 Q7PPI4 A0A1B0C1B4 A0A3F2YSQ9 A0A1A9Y1H6 A0A182VDJ0 A0A182THX6 A0A182WYX7 A0A182LDV4 A0A1S4GN76 A0A182PEN3 A0A3F2YSN9
A0A212EQK9 A0A194PFI0 A0A194R384 A0A2W1BHG8 A0A2A4JUW4 A0A2A4JU52 A0A069DWH7 A0A146LQK9 A0A146MEM0 A0A2H8TMM4 T1HU77 A0A2S2PQU7 A0A2S2PMX6 A0A1B6DBA5 J9KES3 A0A1B6CRS2 A0A2S2QZ54 X2D4J3 A0A2J7Q3G4 D6WED3 B6CY69 A0A1B6LCC7 A0A1B6F5R4 A0A1Y1MI14 A0A1Y1MIE3 A0A1Y1ML21 A0A1B6IXX6 A0A1B6GDB9 N6U4N6 A0A336MYS6 A0A1L8DJV8 A0A1L8DJV5 A0A034VAZ8 A0A336L0X2 A0A0L0CIV0 A0A0R1E314 B4PJW1 A0A1I8PP83 B4MMQ9 B4KZ22 A0A1I8MQX4 B4J247 A0A1B0EY28 A0A0Q5UJS4 Q8IQQ6 C7LAC7 B3NHW0 A0A0Q9XPD1 Q9VVG3 A0A0J9UN26 B5DQ44 A0A3B0KG05 Q9U975 Q8IQQ7 A0A0M3QWZ4 A0A0P8YGX5 A0A0J9RYC3 A0A1I8PP26 C7LA83 A0A336LX44 B4QNU2 B4HKR7 B3M6C3 A0A1A9VYS9 A0A1W4W4W9 A0A1B0C4T3 A0A336KAS2 A0A1W4W4B2 A0A1B0FAG9 A0A1A9WEB0 A0A0K8TQT5 A0A182N207 B4H6L6 A0A084WCY0 Q7PPI4 A0A1B0C1B4 A0A3F2YSQ9 A0A1A9Y1H6 A0A182VDJ0 A0A182THX6 A0A182WYX7 A0A182LDV4 A0A1S4GN76 A0A182PEN3 A0A3F2YSN9
Pubmed
19121390
26354079
22118469
28756777
26334808
26823975
+ More
18362917 19820115 18621706 28004739 23537049 25348373 26108605 17994087 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 22936249 15632085 10433833 26369729 24438588 12364791 20966253
18362917 19820115 18621706 28004739 23537049 25348373 26108605 17994087 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 22936249 15632085 10433833 26369729 24438588 12364791 20966253
EMBL
BABH01005368
BABH01005369
BABH01005370
NWSH01000927
PCG73520.1
ODYU01003081
+ More
SOQ41391.1 KQ459603 KPI92908.1 KQ461194 KPJ06924.1 ODYU01011147 SOQ56711.1 AGBW02013269 OWR43759.1 KQ459605 KPI92042.1 KQ460953 KPJ10306.1 KZ150203 PZC72266.1 NWSH01000575 PCG75556.1 PCG75557.1 GBGD01000837 JAC88052.1 GDHC01009060 JAQ09569.1 GDHC01001187 JAQ17442.1 GFXV01003570 MBW15375.1 ACPB03000713 GGMR01019223 MBY31842.1 GGMR01018115 MBY30734.1 GEDC01014376 JAS22922.1 ABLF02019274 ABLF02019277 ABLF02019278 ABLF02019280 ABLF02019281 ABLF02019283 GEDC01021174 JAS16124.1 GGMS01013823 MBY83026.1 KC713794 AHH29252.1 NEVH01019067 PNF23126.1 KQ971318 EFA00393.2 EU597677 ACH86322.1 GEBQ01025115 GEBQ01018628 JAT14862.1 JAT21349.1 GECZ01024254 JAS45515.1 GEZM01030681 JAV85429.1 GEZM01030679 JAV85431.1 GEZM01030680 JAV85430.1 GECU01015915 JAS91791.1 GECZ01009466 JAS60303.1 APGK01049566 APGK01049567 KB741156 KB632169 ENN73547.1 ERL89466.1 UFQS01002456 UFQT01002456 SSX14090.1 SSX33507.1 GFDF01007342 JAV06742.1 GFDF01007341 JAV06743.1 GAKP01020255 GAKP01020254 JAC38698.1 UFQS01001441 UFQT01001441 SSX10915.1 SSX30593.1 JRES01000428 KNC31379.1 CM000159 KRK02018.1 EDW94730.1 KRK02019.1 CH963847 EDW73465.2 CH933809 EDW17819.1 CH916366 EDV96998.1 AJVK01033670 AJVK01033671 AJVK01033672 CH954178 KQS44048.1 AE014296 BT133104 AAN11709.1 AEX98024.1 AHN58118.1 BT099677 ACV44463.1 EDV51975.2 KQS44049.1 KRG05778.1 KRG05779.1 BT033062 AAF49348.2 ACE82585.1 CM002912 KMZ00221.1 CH379069 EDY73688.2 OUUW01000012 SPP87310.1 AJ243264 CAB53640.1 BT133440 AAN11707.1 AAN11708.1 AFH78572.1 CP012525 ALC44969.1 CH902618 KPU78195.1 KMZ00220.1 KMZ00222.1 BT099633 ACU64822.1 UFQS01000248 UFQT01000248 SSX01993.1 SSX22370.1 CM000363 EDX10844.1 CH480815 EDW41872.1 EDV39743.1 KPU78196.1 JXJN01025645 SSX01994.1 SSX22371.1 CCAG010006264 GDAI01000884 JAI16719.1 CH479214 EDW33444.1 ATLV01022898 KE525337 KFB48074.1 AAAB01008948 EAA10362.5 JXJN01023976 APCN01004624
SOQ41391.1 KQ459603 KPI92908.1 KQ461194 KPJ06924.1 ODYU01011147 SOQ56711.1 AGBW02013269 OWR43759.1 KQ459605 KPI92042.1 KQ460953 KPJ10306.1 KZ150203 PZC72266.1 NWSH01000575 PCG75556.1 PCG75557.1 GBGD01000837 JAC88052.1 GDHC01009060 JAQ09569.1 GDHC01001187 JAQ17442.1 GFXV01003570 MBW15375.1 ACPB03000713 GGMR01019223 MBY31842.1 GGMR01018115 MBY30734.1 GEDC01014376 JAS22922.1 ABLF02019274 ABLF02019277 ABLF02019278 ABLF02019280 ABLF02019281 ABLF02019283 GEDC01021174 JAS16124.1 GGMS01013823 MBY83026.1 KC713794 AHH29252.1 NEVH01019067 PNF23126.1 KQ971318 EFA00393.2 EU597677 ACH86322.1 GEBQ01025115 GEBQ01018628 JAT14862.1 JAT21349.1 GECZ01024254 JAS45515.1 GEZM01030681 JAV85429.1 GEZM01030679 JAV85431.1 GEZM01030680 JAV85430.1 GECU01015915 JAS91791.1 GECZ01009466 JAS60303.1 APGK01049566 APGK01049567 KB741156 KB632169 ENN73547.1 ERL89466.1 UFQS01002456 UFQT01002456 SSX14090.1 SSX33507.1 GFDF01007342 JAV06742.1 GFDF01007341 JAV06743.1 GAKP01020255 GAKP01020254 JAC38698.1 UFQS01001441 UFQT01001441 SSX10915.1 SSX30593.1 JRES01000428 KNC31379.1 CM000159 KRK02018.1 EDW94730.1 KRK02019.1 CH963847 EDW73465.2 CH933809 EDW17819.1 CH916366 EDV96998.1 AJVK01033670 AJVK01033671 AJVK01033672 CH954178 KQS44048.1 AE014296 BT133104 AAN11709.1 AEX98024.1 AHN58118.1 BT099677 ACV44463.1 EDV51975.2 KQS44049.1 KRG05778.1 KRG05779.1 BT033062 AAF49348.2 ACE82585.1 CM002912 KMZ00221.1 CH379069 EDY73688.2 OUUW01000012 SPP87310.1 AJ243264 CAB53640.1 BT133440 AAN11707.1 AAN11708.1 AFH78572.1 CP012525 ALC44969.1 CH902618 KPU78195.1 KMZ00220.1 KMZ00222.1 BT099633 ACU64822.1 UFQS01000248 UFQT01000248 SSX01993.1 SSX22370.1 CM000363 EDX10844.1 CH480815 EDW41872.1 EDV39743.1 KPU78196.1 JXJN01025645 SSX01994.1 SSX22371.1 CCAG010006264 GDAI01000884 JAI16719.1 CH479214 EDW33444.1 ATLV01022898 KE525337 KFB48074.1 AAAB01008948 EAA10362.5 JXJN01023976 APCN01004624
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000015103
+ More
UP000007819 UP000235965 UP000007266 UP000019118 UP000030742 UP000037069 UP000002282 UP000095300 UP000007798 UP000009192 UP000095301 UP000001070 UP000092462 UP000008711 UP000000803 UP000001819 UP000268350 UP000092553 UP000007801 UP000000304 UP000001292 UP000078200 UP000192221 UP000092460 UP000092444 UP000091820 UP000075884 UP000008744 UP000030765 UP000007062 UP000075840 UP000092443 UP000075903 UP000075902 UP000076407 UP000075882 UP000075885
UP000007819 UP000235965 UP000007266 UP000019118 UP000030742 UP000037069 UP000002282 UP000095300 UP000007798 UP000009192 UP000095301 UP000001070 UP000092462 UP000008711 UP000000803 UP000001819 UP000268350 UP000092553 UP000007801 UP000000304 UP000001292 UP000078200 UP000192221 UP000092460 UP000092444 UP000091820 UP000075884 UP000008744 UP000030765 UP000007062 UP000075840 UP000092443 UP000075903 UP000075902 UP000076407 UP000075882 UP000075885
Pfam
PF00209 SNF
SUPFAM
SSF161070
SSF161070
ProteinModelPortal
H9JLX6
A0A2A4JQ39
A0A2H1VKR8
A0A194PIN1
A0A194QNJ1
A0A2H1WUH2
+ More
A0A212EQK9 A0A194PFI0 A0A194R384 A0A2W1BHG8 A0A2A4JUW4 A0A2A4JU52 A0A069DWH7 A0A146LQK9 A0A146MEM0 A0A2H8TMM4 T1HU77 A0A2S2PQU7 A0A2S2PMX6 A0A1B6DBA5 J9KES3 A0A1B6CRS2 A0A2S2QZ54 X2D4J3 A0A2J7Q3G4 D6WED3 B6CY69 A0A1B6LCC7 A0A1B6F5R4 A0A1Y1MI14 A0A1Y1MIE3 A0A1Y1ML21 A0A1B6IXX6 A0A1B6GDB9 N6U4N6 A0A336MYS6 A0A1L8DJV8 A0A1L8DJV5 A0A034VAZ8 A0A336L0X2 A0A0L0CIV0 A0A0R1E314 B4PJW1 A0A1I8PP83 B4MMQ9 B4KZ22 A0A1I8MQX4 B4J247 A0A1B0EY28 A0A0Q5UJS4 Q8IQQ6 C7LAC7 B3NHW0 A0A0Q9XPD1 Q9VVG3 A0A0J9UN26 B5DQ44 A0A3B0KG05 Q9U975 Q8IQQ7 A0A0M3QWZ4 A0A0P8YGX5 A0A0J9RYC3 A0A1I8PP26 C7LA83 A0A336LX44 B4QNU2 B4HKR7 B3M6C3 A0A1A9VYS9 A0A1W4W4W9 A0A1B0C4T3 A0A336KAS2 A0A1W4W4B2 A0A1B0FAG9 A0A1A9WEB0 A0A0K8TQT5 A0A182N207 B4H6L6 A0A084WCY0 Q7PPI4 A0A1B0C1B4 A0A3F2YSQ9 A0A1A9Y1H6 A0A182VDJ0 A0A182THX6 A0A182WYX7 A0A182LDV4 A0A1S4GN76 A0A182PEN3 A0A3F2YSN9
A0A212EQK9 A0A194PFI0 A0A194R384 A0A2W1BHG8 A0A2A4JUW4 A0A2A4JU52 A0A069DWH7 A0A146LQK9 A0A146MEM0 A0A2H8TMM4 T1HU77 A0A2S2PQU7 A0A2S2PMX6 A0A1B6DBA5 J9KES3 A0A1B6CRS2 A0A2S2QZ54 X2D4J3 A0A2J7Q3G4 D6WED3 B6CY69 A0A1B6LCC7 A0A1B6F5R4 A0A1Y1MI14 A0A1Y1MIE3 A0A1Y1ML21 A0A1B6IXX6 A0A1B6GDB9 N6U4N6 A0A336MYS6 A0A1L8DJV8 A0A1L8DJV5 A0A034VAZ8 A0A336L0X2 A0A0L0CIV0 A0A0R1E314 B4PJW1 A0A1I8PP83 B4MMQ9 B4KZ22 A0A1I8MQX4 B4J247 A0A1B0EY28 A0A0Q5UJS4 Q8IQQ6 C7LAC7 B3NHW0 A0A0Q9XPD1 Q9VVG3 A0A0J9UN26 B5DQ44 A0A3B0KG05 Q9U975 Q8IQQ7 A0A0M3QWZ4 A0A0P8YGX5 A0A0J9RYC3 A0A1I8PP26 C7LA83 A0A336LX44 B4QNU2 B4HKR7 B3M6C3 A0A1A9VYS9 A0A1W4W4W9 A0A1B0C4T3 A0A336KAS2 A0A1W4W4B2 A0A1B0FAG9 A0A1A9WEB0 A0A0K8TQT5 A0A182N207 B4H6L6 A0A084WCY0 Q7PPI4 A0A1B0C1B4 A0A3F2YSQ9 A0A1A9Y1H6 A0A182VDJ0 A0A182THX6 A0A182WYX7 A0A182LDV4 A0A1S4GN76 A0A182PEN3 A0A3F2YSN9
PDB
4XPT
E-value=2.22594e-10,
Score=160
Ontologies
GO
PANTHER
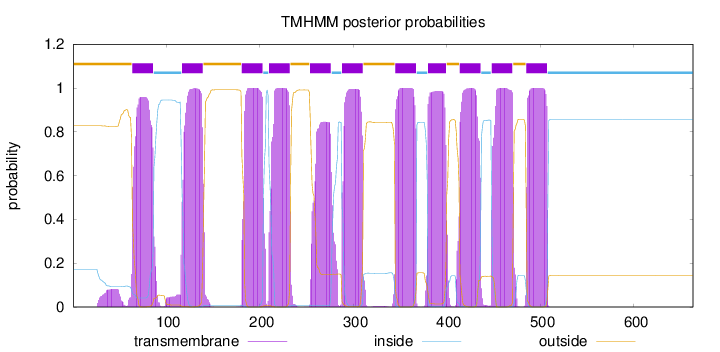
Topology
Length:
663
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
242.06869
Exp number, first 60 AAs:
1.80476
Total prob of N-in:
0.17256
outside
1 - 63
TMhelix
64 - 86
inside
87 - 116
TMhelix
117 - 139
outside
140 - 180
TMhelix
181 - 203
inside
204 - 209
TMhelix
210 - 232
outside
233 - 253
TMhelix
254 - 276
inside
277 - 287
TMhelix
288 - 310
outside
311 - 344
TMhelix
345 - 367
inside
368 - 379
TMhelix
380 - 399
outside
400 - 413
TMhelix
414 - 436
inside
437 - 447
TMhelix
448 - 470
outside
471 - 484
TMhelix
485 - 507
inside
508 - 663
Population Genetic Test Statistics
Pi
205.936711
Theta
16.595667
Tajima's D
0.565641
CLR
0.285575
CSRT
0.543872806359682
Interpretation
Uncertain
