Pre Gene Modal
BGIBMGA010527
Annotation
PREDICTED:_endoplasmic_reticulum_lectin_1_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.949
Sequence
CDS
ATGAGAGTGAAAATTGTTATGTTAAGTTTAGTTTATAGCTGCATAGGTATTGAACATGACTTTAAAGGATTCGATGATAGCATTTTGTTTGGGATAAATTGGCCTGGTAATCCCAATTTAAACGAAAATTTTGTTGATGGAGAAAAAAACAAGTCTCAGAACCAAGAGTTAGTTAAAGTAACAACAACAAATAAAGAGAGTTACGAATGTCAACTGCCGGAACTTCGTAGTACGGAGTCTACCAGCATTGATGATTACGATGGTCCATCACCGCTTCACCTTCTAAAGCCTTTGATGAACAAAGAAATGTGTTCATACAGATTGGAAAGCTATTGGAGCTATGAAGTATGCCATGGAAGGTATATCCGGCAGTATCACGAAGAAAGAGAGGGCAAACAAATCAACACCCAAGAATATTTCTTGGGATATTGGAGCTCTGAAAAGCAGGCTAAGCTGGAGGCTGAAATGAAAGCGGCACAGGAATCCAAACAAATCCCCAAGACCACCAAAGTAGAGGGAGTTGCTCTACCTAACGTTGAAATAGTAATGGATGATGGTACAATTTGTGATTTAAATGGCAAACCACGACTAACTAGGGTTCATTATGTGTGTTACACACATGGAAAGCATGAGGTTTATTCCTTCAGAGAGACTTCTACTTGCGAGTACGAGATCATTATTCTGTCGCCGTTTCTATGTGAACATCCACAGTTCAAACCTATGGATGTTAGTGAGAATGTTATAAACTGCCTCCCAATCGGAGACGCTCCAAGGAAGCCAAGGGACTTATTGAAAATGGAAGCGAACAGTTTGAGATTTCATCATCAGAGCATAAGACTCATGGATAATGCTGACCAGACAGTTAAGGATATACTTGCAGTTTTGAAAGTCGGAAAATTTGAGAAGGACGGCGAAACCCATTTGAAATTCGAACTGCACCCGCTGGGTGACGACGAGGATATGACGTCAGACCCACCGAAGCCAGTACCGAGCGTCGACAAGCGCCCTCCGCCAATCACGGATGATTCTCCCGTACGAGCCTTCCTTAACGGGGAAAATTGTCTAAATGGTGGTACAGGCTGGTGGAAATACGAGTTCTGCTACGGGAAGCACGTGCTCCAGTACCACATAGATCGCGCCGGCGAGAAGACCACCGTAATGTTGGGTCGATTTGATGAGAAAGCGCATTTGGACTGGATCAAGGATAATAAAGGGAAAGCTCCGAAACCTATAGATCAAAGAATTTCAATATCGCATTTCTATTCGAACGGCGACGTTTGCGACAAAACCGGCCGACCCCGGCAGACCGAGGTCAAACTTAAATGTCTAGAGAATTCGTCGAGTCCAGCTCAAGTCTCCCTGTACTTGTTGGAACCTAAAACTTGTCATTACATACTGGGCGTGGAATCGCCTTTGATATGCGACATATTGCCTTTGGCCGACGAGAATGGCCTTATAAAAACTGTGAGAGAGGCCTTGGAAAAGAAGAAGGAGAGGAGTGAGGCTGTCGATGATGTCTCAGACAAGAATGTACCAAAATACGAGAATGATTAG
Protein
MRVKIVMLSLVYSCIGIEHDFKGFDDSILFGINWPGNPNLNENFVDGEKNKSQNQELVKVTTTNKESYECQLPELRSTESTSIDDYDGPSPLHLLKPLMNKEMCSYRLESYWSYEVCHGRYIRQYHEEREGKQINTQEYFLGYWSSEKQAKLEAEMKAAQESKQIPKTTKVEGVALPNVEIVMDDGTICDLNGKPRLTRVHYVCYTHGKHEVYSFRETSTCEYEIIILSPFLCEHPQFKPMDVSENVINCLPIGDAPRKPRDLLKMEANSLRFHHQSIRLMDNADQTVKDILAVLKVGKFEKDGETHLKFELHPLGDDEDMTSDPPKPVPSVDKRPPPITDDSPVRAFLNGENCLNGGTGWWKYEFCYGKHVLQYHIDRAGEKTTVMLGRFDEKAHLDWIKDNKGKAPKPIDQRISISHFYSNGDVCDKTGRPRQTEVKLKCLENSSSPAQVSLYLLEPKTCHYILGVESPLICDILPLADENGLIKTVREALEKKKERSEAVDDVSDKNVPKYEND
Summary
Uniprot
H9JLX5
A0A2A4IUE4
A0A1E1W6W4
A0A2H1UZM0
A0A194PIN6
A0A194QNI6
+ More
A0A212FA96 S4P2C8 A0A2J7Q8A5 A0A2J7Q8B1 A0A158NR90 A0A067RK67 A0A195BE96 A0A151X522 F4WB20 A0A1B6G9A4 A0A1Y1LS51 A0A1B6LLQ5 A0A195FJL0 A0A310SX19 A0A0M9A1L8 E2BLR9 A0A026W5R4 D6WC47 A0A1W4X4H6 A0A0L7RE36 A0A0J7L920 A0A151IFT5 A0A1B6HPD9 A0A151IU40 E2A0E9 A0A232FD42 E0VNV4 E9IRY1 A0A1J1HJB0 A0A1L8DW66 A0A1L8DW80 A0A2A3EF46 A0A2P8Y8I4 A0A336LLP1 T1DGN7 A0A336LQN8 A0A182NE86 A0A182Q643 J3JXL5 N6UNI9 A0A182IZ96 A0A084VP97 A0A182PB49 A0A154PB30 A0A023EW34 A0A1Q3FH37 A0A182VV26 A0A182K168 U5EP20 Q7QG77 Q170H4 A0A1S4FI03 A0A182VAR6 A0A182GN56 A0A182L5S3 A0A182UBJ5 A0A182XCN3 Q170H5 A0A182RPP9 A0A182IE07 A0A224XEI7 A0A1Q3FH53 A0A182Y5R2 B0X6J9 A0A182LWM3 T1DJG7 A0A0P4VUT3 A0A087ZYR0 A0A2M4BLY1 W5J6P8 A0A2M4BL55 A0A2M4BL41 A0A2M3Z3D9 A0A2M3Z398 A0A2M3Z3B5 A0A2K8JM33 A0A2M4A6S9 T1I6M7 A0A0P5WAY6 A0A0P6JAY5 A0A0P5XI52 A0A182F4E8 J9K540 E9HAB7 A0A146LN24 A0A0A9X029 A0A1D2NAS4 A0A2S2PSH0 A0A0P6EQK3 A0A0P6F725 A0A0P6BFQ2 A0A0N8C1M2 A0A3S3P692 X1WUR7
A0A212FA96 S4P2C8 A0A2J7Q8A5 A0A2J7Q8B1 A0A158NR90 A0A067RK67 A0A195BE96 A0A151X522 F4WB20 A0A1B6G9A4 A0A1Y1LS51 A0A1B6LLQ5 A0A195FJL0 A0A310SX19 A0A0M9A1L8 E2BLR9 A0A026W5R4 D6WC47 A0A1W4X4H6 A0A0L7RE36 A0A0J7L920 A0A151IFT5 A0A1B6HPD9 A0A151IU40 E2A0E9 A0A232FD42 E0VNV4 E9IRY1 A0A1J1HJB0 A0A1L8DW66 A0A1L8DW80 A0A2A3EF46 A0A2P8Y8I4 A0A336LLP1 T1DGN7 A0A336LQN8 A0A182NE86 A0A182Q643 J3JXL5 N6UNI9 A0A182IZ96 A0A084VP97 A0A182PB49 A0A154PB30 A0A023EW34 A0A1Q3FH37 A0A182VV26 A0A182K168 U5EP20 Q7QG77 Q170H4 A0A1S4FI03 A0A182VAR6 A0A182GN56 A0A182L5S3 A0A182UBJ5 A0A182XCN3 Q170H5 A0A182RPP9 A0A182IE07 A0A224XEI7 A0A1Q3FH53 A0A182Y5R2 B0X6J9 A0A182LWM3 T1DJG7 A0A0P4VUT3 A0A087ZYR0 A0A2M4BLY1 W5J6P8 A0A2M4BL55 A0A2M4BL41 A0A2M3Z3D9 A0A2M3Z398 A0A2M3Z3B5 A0A2K8JM33 A0A2M4A6S9 T1I6M7 A0A0P5WAY6 A0A0P6JAY5 A0A0P5XI52 A0A182F4E8 J9K540 E9HAB7 A0A146LN24 A0A0A9X029 A0A1D2NAS4 A0A2S2PSH0 A0A0P6EQK3 A0A0P6F725 A0A0P6BFQ2 A0A0N8C1M2 A0A3S3P692 X1WUR7
Pubmed
19121390
26354079
22118469
23622113
21347285
24845553
+ More
21719571 28004739 20798317 24508170 30249741 18362917 19820115 28648823 20566863 21282665 29403074 24330624 22516182 23537049 24438588 24945155 12364791 17510324 26483478 20966253 25244985 27129103 20920257 23761445 21292972 26823975 25401762 27289101
21719571 28004739 20798317 24508170 30249741 18362917 19820115 28648823 20566863 21282665 29403074 24330624 22516182 23537049 24438588 24945155 12364791 17510324 26483478 20966253 25244985 27129103 20920257 23761445 21292972 26823975 25401762 27289101
EMBL
BABH01005335
BABH01005336
NWSH01007909
PCG62760.1
GDQN01008360
JAT82694.1
+ More
ODYU01000002 SOQ34047.1 KQ459603 KPI92913.1 KQ461194 KPJ06919.1 AGBW02009509 OWR50648.1 GAIX01012085 JAA80475.1 NEVH01016966 PNF24821.1 PNF24823.1 ADTU01023825 KK852424 KDR24187.1 KQ976504 KYM82893.1 KQ982540 KYQ55424.1 GL888056 EGI68640.1 GECZ01022859 GECZ01010759 JAS46910.1 JAS59010.1 GEZM01050228 JAV75668.1 GEBQ01015380 JAT24597.1 KQ981522 KYN40588.1 KQ759870 OAD62414.1 KQ435765 KOX75337.1 GL449036 EFN83403.1 KK107390 QOIP01000007 EZA51410.1 RLU20262.1 KQ971316 EEZ97851.1 KQ414613 KOC69075.1 LBMM01000197 KMR04520.1 KQ977771 KYN00008.1 GECU01031154 JAS76552.1 KQ980970 KYN10927.1 GL435586 EFN73086.1 NNAY01000440 OXU28378.1 DS235354 EEB15060.1 GL765283 EFZ16592.1 CVRI01000001 CRK86361.1 GFDF01003408 JAV10676.1 GFDF01003377 JAV10707.1 KZ288269 PBC29902.1 PYGN01000806 PSN40562.1 UFQT01000048 SSX18972.1 GALA01000127 JAA94725.1 SSX18973.1 AXCN02000866 BT127984 KB632093 AEE62946.1 ERL88683.1 APGK01004410 APGK01027778 KB740671 KB735736 ENN79624.1 ENN83315.1 ATLV01014987 KE524999 KFB39791.1 KQ434864 KZC09119.1 GAPW01001039 JAC12559.1 GFDL01008178 JAV26867.1 GANO01000353 JAB59518.1 AAAB01008839 EAA05924.4 CH477473 EAT40367.1 JXUM01014438 JXUM01014439 KQ560403 KXJ82588.1 EAT40368.1 APCN01004546 GFTR01007012 JAW09414.1 GFDL01008180 JAV26865.1 DS232416 EDS41482.1 AXCM01002140 GALA01000276 JAA94576.1 GDKW01000802 JAI55793.1 GGFJ01004647 MBW53788.1 ADMH02002133 ETN58444.1 GGFJ01004644 MBW53785.1 GGFJ01004646 MBW53787.1 GGFM01002290 MBW23041.1 GGFM01002239 MBW22990.1 GGFM01002244 MBW22995.1 KY031135 ATU82886.1 GGFK01003158 MBW36479.1 ACPB03010942 GDIP01088653 LRGB01003024 JAM15062.1 KZS05002.1 GDIQ01020226 JAN74511.1 GDIP01071747 JAM31968.1 ABLF02030293 GL732611 EFX71320.1 GDHC01010869 JAQ07760.1 GBHO01030240 GBRD01012846 JAG13364.1 JAG52980.1 LJIJ01000115 ODN02347.1 GGMR01019803 MBY32422.1 GDIQ01059768 JAN34969.1 GDIQ01052370 JAN42367.1 GDIP01015333 JAM88382.1 GDIQ01123683 JAL28043.1 NCKU01000359 RWS15814.1 ABLF02009754
ODYU01000002 SOQ34047.1 KQ459603 KPI92913.1 KQ461194 KPJ06919.1 AGBW02009509 OWR50648.1 GAIX01012085 JAA80475.1 NEVH01016966 PNF24821.1 PNF24823.1 ADTU01023825 KK852424 KDR24187.1 KQ976504 KYM82893.1 KQ982540 KYQ55424.1 GL888056 EGI68640.1 GECZ01022859 GECZ01010759 JAS46910.1 JAS59010.1 GEZM01050228 JAV75668.1 GEBQ01015380 JAT24597.1 KQ981522 KYN40588.1 KQ759870 OAD62414.1 KQ435765 KOX75337.1 GL449036 EFN83403.1 KK107390 QOIP01000007 EZA51410.1 RLU20262.1 KQ971316 EEZ97851.1 KQ414613 KOC69075.1 LBMM01000197 KMR04520.1 KQ977771 KYN00008.1 GECU01031154 JAS76552.1 KQ980970 KYN10927.1 GL435586 EFN73086.1 NNAY01000440 OXU28378.1 DS235354 EEB15060.1 GL765283 EFZ16592.1 CVRI01000001 CRK86361.1 GFDF01003408 JAV10676.1 GFDF01003377 JAV10707.1 KZ288269 PBC29902.1 PYGN01000806 PSN40562.1 UFQT01000048 SSX18972.1 GALA01000127 JAA94725.1 SSX18973.1 AXCN02000866 BT127984 KB632093 AEE62946.1 ERL88683.1 APGK01004410 APGK01027778 KB740671 KB735736 ENN79624.1 ENN83315.1 ATLV01014987 KE524999 KFB39791.1 KQ434864 KZC09119.1 GAPW01001039 JAC12559.1 GFDL01008178 JAV26867.1 GANO01000353 JAB59518.1 AAAB01008839 EAA05924.4 CH477473 EAT40367.1 JXUM01014438 JXUM01014439 KQ560403 KXJ82588.1 EAT40368.1 APCN01004546 GFTR01007012 JAW09414.1 GFDL01008180 JAV26865.1 DS232416 EDS41482.1 AXCM01002140 GALA01000276 JAA94576.1 GDKW01000802 JAI55793.1 GGFJ01004647 MBW53788.1 ADMH02002133 ETN58444.1 GGFJ01004644 MBW53785.1 GGFJ01004646 MBW53787.1 GGFM01002290 MBW23041.1 GGFM01002239 MBW22990.1 GGFM01002244 MBW22995.1 KY031135 ATU82886.1 GGFK01003158 MBW36479.1 ACPB03010942 GDIP01088653 LRGB01003024 JAM15062.1 KZS05002.1 GDIQ01020226 JAN74511.1 GDIP01071747 JAM31968.1 ABLF02030293 GL732611 EFX71320.1 GDHC01010869 JAQ07760.1 GBHO01030240 GBRD01012846 JAG13364.1 JAG52980.1 LJIJ01000115 ODN02347.1 GGMR01019803 MBY32422.1 GDIQ01059768 JAN34969.1 GDIQ01052370 JAN42367.1 GDIP01015333 JAM88382.1 GDIQ01123683 JAL28043.1 NCKU01000359 RWS15814.1 ABLF02009754
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000235965
+ More
UP000005205 UP000027135 UP000078540 UP000075809 UP000007755 UP000078541 UP000053105 UP000008237 UP000053097 UP000279307 UP000007266 UP000192223 UP000053825 UP000036403 UP000078542 UP000078492 UP000000311 UP000215335 UP000009046 UP000183832 UP000242457 UP000245037 UP000075884 UP000075886 UP000030742 UP000019118 UP000075880 UP000030765 UP000075885 UP000076502 UP000075920 UP000075881 UP000007062 UP000008820 UP000075903 UP000069940 UP000249989 UP000075882 UP000075902 UP000076407 UP000075900 UP000075840 UP000076408 UP000002320 UP000075883 UP000005203 UP000000673 UP000015103 UP000076858 UP000069272 UP000007819 UP000000305 UP000094527 UP000285301
UP000005205 UP000027135 UP000078540 UP000075809 UP000007755 UP000078541 UP000053105 UP000008237 UP000053097 UP000279307 UP000007266 UP000192223 UP000053825 UP000036403 UP000078542 UP000078492 UP000000311 UP000215335 UP000009046 UP000183832 UP000242457 UP000245037 UP000075884 UP000075886 UP000030742 UP000019118 UP000075880 UP000030765 UP000075885 UP000076502 UP000075920 UP000075881 UP000007062 UP000008820 UP000075903 UP000069940 UP000249989 UP000075882 UP000075902 UP000076407 UP000075900 UP000075840 UP000076408 UP000002320 UP000075883 UP000005203 UP000000673 UP000015103 UP000076858 UP000069272 UP000007819 UP000000305 UP000094527 UP000285301
PRIDE
Pfam
PF07915 PRKCSH
Gene 3D
ProteinModelPortal
H9JLX5
A0A2A4IUE4
A0A1E1W6W4
A0A2H1UZM0
A0A194PIN6
A0A194QNI6
+ More
A0A212FA96 S4P2C8 A0A2J7Q8A5 A0A2J7Q8B1 A0A158NR90 A0A067RK67 A0A195BE96 A0A151X522 F4WB20 A0A1B6G9A4 A0A1Y1LS51 A0A1B6LLQ5 A0A195FJL0 A0A310SX19 A0A0M9A1L8 E2BLR9 A0A026W5R4 D6WC47 A0A1W4X4H6 A0A0L7RE36 A0A0J7L920 A0A151IFT5 A0A1B6HPD9 A0A151IU40 E2A0E9 A0A232FD42 E0VNV4 E9IRY1 A0A1J1HJB0 A0A1L8DW66 A0A1L8DW80 A0A2A3EF46 A0A2P8Y8I4 A0A336LLP1 T1DGN7 A0A336LQN8 A0A182NE86 A0A182Q643 J3JXL5 N6UNI9 A0A182IZ96 A0A084VP97 A0A182PB49 A0A154PB30 A0A023EW34 A0A1Q3FH37 A0A182VV26 A0A182K168 U5EP20 Q7QG77 Q170H4 A0A1S4FI03 A0A182VAR6 A0A182GN56 A0A182L5S3 A0A182UBJ5 A0A182XCN3 Q170H5 A0A182RPP9 A0A182IE07 A0A224XEI7 A0A1Q3FH53 A0A182Y5R2 B0X6J9 A0A182LWM3 T1DJG7 A0A0P4VUT3 A0A087ZYR0 A0A2M4BLY1 W5J6P8 A0A2M4BL55 A0A2M4BL41 A0A2M3Z3D9 A0A2M3Z398 A0A2M3Z3B5 A0A2K8JM33 A0A2M4A6S9 T1I6M7 A0A0P5WAY6 A0A0P6JAY5 A0A0P5XI52 A0A182F4E8 J9K540 E9HAB7 A0A146LN24 A0A0A9X029 A0A1D2NAS4 A0A2S2PSH0 A0A0P6EQK3 A0A0P6F725 A0A0P6BFQ2 A0A0N8C1M2 A0A3S3P692 X1WUR7
A0A212FA96 S4P2C8 A0A2J7Q8A5 A0A2J7Q8B1 A0A158NR90 A0A067RK67 A0A195BE96 A0A151X522 F4WB20 A0A1B6G9A4 A0A1Y1LS51 A0A1B6LLQ5 A0A195FJL0 A0A310SX19 A0A0M9A1L8 E2BLR9 A0A026W5R4 D6WC47 A0A1W4X4H6 A0A0L7RE36 A0A0J7L920 A0A151IFT5 A0A1B6HPD9 A0A151IU40 E2A0E9 A0A232FD42 E0VNV4 E9IRY1 A0A1J1HJB0 A0A1L8DW66 A0A1L8DW80 A0A2A3EF46 A0A2P8Y8I4 A0A336LLP1 T1DGN7 A0A336LQN8 A0A182NE86 A0A182Q643 J3JXL5 N6UNI9 A0A182IZ96 A0A084VP97 A0A182PB49 A0A154PB30 A0A023EW34 A0A1Q3FH37 A0A182VV26 A0A182K168 U5EP20 Q7QG77 Q170H4 A0A1S4FI03 A0A182VAR6 A0A182GN56 A0A182L5S3 A0A182UBJ5 A0A182XCN3 Q170H5 A0A182RPP9 A0A182IE07 A0A224XEI7 A0A1Q3FH53 A0A182Y5R2 B0X6J9 A0A182LWM3 T1DJG7 A0A0P4VUT3 A0A087ZYR0 A0A2M4BLY1 W5J6P8 A0A2M4BL55 A0A2M4BL41 A0A2M3Z3D9 A0A2M3Z398 A0A2M3Z3B5 A0A2K8JM33 A0A2M4A6S9 T1I6M7 A0A0P5WAY6 A0A0P6JAY5 A0A0P5XI52 A0A182F4E8 J9K540 E9HAB7 A0A146LN24 A0A0A9X029 A0A1D2NAS4 A0A2S2PSH0 A0A0P6EQK3 A0A0P6F725 A0A0P6BFQ2 A0A0N8C1M2 A0A3S3P692 X1WUR7
PDB
3AIH
E-value=4.60018e-16,
Score=208
Ontologies
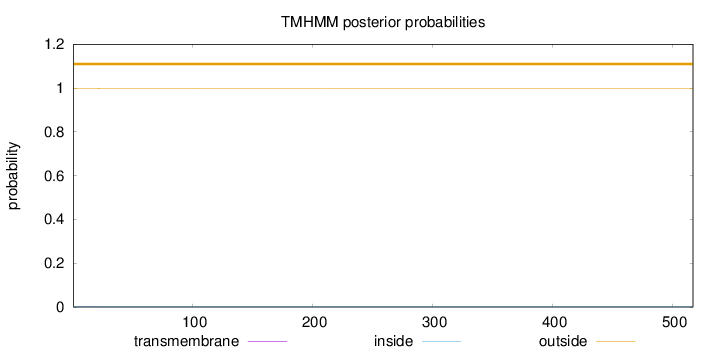
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.942139

Length:
517
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01275
Exp number, first 60 AAs:
0.01233
Total prob of N-in:
0.00203
outside
1 - 517
Population Genetic Test Statistics
Pi
18.28602
Theta
41.10047
Tajima's D
-1.791155
CLR
210.637838
CSRT
0.0285985700714964
Interpretation
Uncertain
