Gene
KWMTBOMO07081
Annotation
Cornichon_[Operophtera_brumata]
Full name
Protein cornichon
Location in the cell
PlasmaMembrane Reliability : 4.873
Sequence
CDS
ATGGCGTTTAGTTTTCCAGCCTTTGCGTATATTGTTGCCTTAATAACCGATGCATTTTTAATATTTTTCTCTATATTTCACGTTATAGCCTTCGACGAACTAAAAACTGACTACAAAAATCCCATCGATCAATGCAATAGTTTGAATCCTTTGGTTTTGCCGGAATATTTACTGCATTTGCTCATTAATGTACTGTTCTTGCTGTCCGGAGAATGGTTTTCGTTATTGATCAACATTCCTTTAATACTATACCACATACACAGATACTACACAAGGCCAGTCATGTCAGGTCCAGGTCTGTATGACCCGACAAGTATTATGAATGCAGATGTATTAACTTCATGTCAGAGGGAAGGCTGGATAAAGCTGGCATTTTACTTGCTGTCGTTCTTCCTATATCTTTATGGAATGATCGTGGTGTTGATTGCGGCGTAA
Protein
MAFSFPAFAYIVALITDAFLIFFSIFHVIAFDELKTDYKNPIDQCNSLNPLVLPEYLLHLLINVLFLLSGEWFSLLINIPLILYHIHRYYTRPVMSGPGLYDPTSIMNADVLTSCQREGWIKLAFYLLSFFLYLYGMIVVLIAA
Summary
Description
Acts as cargo receptor necessary for the transportation of gurken (grk) to a transitional endoplasmic reticulum (tER) site and promotes its incorporation into coat protein complex II (COPII) vesicles. Associated with gurken, produces a signal received by torpedo resulting in a signaling pathway that first establishes posterior follicle cell fates and normal localization of the anterior and posterior determinants, later they act in a signaling event inducing dorsal follicle cell fates and regulating the dorsal-ventral pattern of egg and embryo.
Acts as cargo receptor necessary for the transportation of gurken (grk) to a transitional endoplasmic reticulum (tER) site and promotes its incorporation into coat protein complex II (COPII) vesicles. Associated with gurken, produces a signal received by torpedo resulting in a signaling pathway that first establishes posterior follicle cell fates and normal localization of the anterior and posterior determinants, later they act in a signaling event inducing dorsal follicle cell fates and regulating the dorsal-ventral pattern of egg and embryo (By similarity).
Acts as cargo receptor necessary for the transportation of gurken (grk) to a transitional endoplasmic reticulum (tER) site and promotes its incorporation into coat protein complex II (COPII) vesicles. Associated with gurken, produces a signal received by torpedo resulting in a signaling pathway that first establishes posterior follicle cell fates and normal localization of the anterior and posterior determinants, later they act in a signaling event inducing dorsal follicle cell fates and regulating the dorsal-ventral pattern of egg and embryo (By similarity).
Subunit
Interacts with grk.
Similarity
Belongs to the cornichon family.
Keywords
Complete proteome
Developmental protein
Endoplasmic reticulum
ER-Golgi transport
Membrane
Protein transport
Receptor
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Protein cornichon
Uniprot
A0A0L7LM61
A0A3S2TPT5
A0A2H1UZK1
A0A212FA84
S4PS54
D6WLE9
+ More
I4DP33 A0A182JBA0 A0A1L8DV30 U5EM43 A0A1B0GPQ2 A0A2P8ZLZ4 A0A139WH69 A0A2J7R8M6 A0A067RD48 A0A2M3ZE16 A0A2M4AVF8 W5JVX0 A0A182F316 R4WDC1 A0A194PJL3 Q172M8 A0A023EE82 A0A1Q3FBV4 B0WRE4 A0A182NUP5 A0A0T6B2P7 A0A182RZB4 A0A182WGB7 A0A1B6L7H4 A0A023F9P2 R4G4D4 A0A182UXN3 A0A182WXL8 Q7PWG7 A0A182I7I2 A0A182QLC5 A0A224XVW3 A0A084WQK5 A0A182HCJ5 E9HJT3 A0A1W4WSK6 A0A2R7W0T9 A0A0P5VVU2 A0A164YC13 A0A2P2HYL8 A0A182Y0H4 A0A182KF06 A0A182T2V6 A0A182PVU2 A0A182LMJ8 K7J0R5 A0A226EKL1 A0A0A9YLW8 A0A336MAR6 A0A0P5IDQ6 J3JU84 A0A0P6G364 A0A1D2MI40 E0VNZ1 A0A0N7ZD36 A0A3B0KCW3 Q29NM5 B4GQN2 A0A0L0BZF8 A0A1I8MH61 A0A1I8Q4B7 A0A0C9RR36 B4Q701 A0A1W4VWF9 B3MJN8 A0A0P5YS29 B4I217 M9PDJ6 P49858 A0A1L8EHC4 A0A0M5J413 B4MU61 B4NXP4 B3N6D0 D3TRN8 A0A0K8U5G9 A0A087UX33 A0A034W1T8 A0A2S2N644 A0A0L7RDL0 A0A0Q9WAH1 B4KI22 P52159 C1BZZ6 A0A1A9WM33 J9JJ49 A0A0Q9X5X3 A0A0P6ENN8 T1K2W9 A0A2H8TJM0 C1BUM3 A0A0N8DPJ5 K1QF47 R7TQB5
I4DP33 A0A182JBA0 A0A1L8DV30 U5EM43 A0A1B0GPQ2 A0A2P8ZLZ4 A0A139WH69 A0A2J7R8M6 A0A067RD48 A0A2M3ZE16 A0A2M4AVF8 W5JVX0 A0A182F316 R4WDC1 A0A194PJL3 Q172M8 A0A023EE82 A0A1Q3FBV4 B0WRE4 A0A182NUP5 A0A0T6B2P7 A0A182RZB4 A0A182WGB7 A0A1B6L7H4 A0A023F9P2 R4G4D4 A0A182UXN3 A0A182WXL8 Q7PWG7 A0A182I7I2 A0A182QLC5 A0A224XVW3 A0A084WQK5 A0A182HCJ5 E9HJT3 A0A1W4WSK6 A0A2R7W0T9 A0A0P5VVU2 A0A164YC13 A0A2P2HYL8 A0A182Y0H4 A0A182KF06 A0A182T2V6 A0A182PVU2 A0A182LMJ8 K7J0R5 A0A226EKL1 A0A0A9YLW8 A0A336MAR6 A0A0P5IDQ6 J3JU84 A0A0P6G364 A0A1D2MI40 E0VNZ1 A0A0N7ZD36 A0A3B0KCW3 Q29NM5 B4GQN2 A0A0L0BZF8 A0A1I8MH61 A0A1I8Q4B7 A0A0C9RR36 B4Q701 A0A1W4VWF9 B3MJN8 A0A0P5YS29 B4I217 M9PDJ6 P49858 A0A1L8EHC4 A0A0M5J413 B4MU61 B4NXP4 B3N6D0 D3TRN8 A0A0K8U5G9 A0A087UX33 A0A034W1T8 A0A2S2N644 A0A0L7RDL0 A0A0Q9WAH1 B4KI22 P52159 C1BZZ6 A0A1A9WM33 J9JJ49 A0A0Q9X5X3 A0A0P6ENN8 T1K2W9 A0A2H8TJM0 C1BUM3 A0A0N8DPJ5 K1QF47 R7TQB5
Pubmed
26227816
22118469
23622113
18362917
19820115
22651552
+ More
29403074 24845553 20920257 23761445 23691247 26354079 17510324 26999592 24945155 25474469 12364791 14747013 17210077 24438588 26483478 21292972 25244985 20966253 20075255 25401762 26823975 22516182 23537049 27289101 20566863 15632085 17994087 26108605 25315136 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7540118 10471707 16396907 17550304 20353571 25348373 22992520 23254933
29403074 24845553 20920257 23761445 23691247 26354079 17510324 26999592 24945155 25474469 12364791 14747013 17210077 24438588 26483478 21292972 25244985 20966253 20075255 25401762 26823975 22516182 23537049 27289101 20566863 15632085 17994087 26108605 25315136 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7540118 10471707 16396907 17550304 20353571 25348373 22992520 23254933
EMBL
JTDY01000662
KOB76296.1
RSAL01000028
RVE51855.1
ODYU01000002
SOQ34048.1
+ More
AGBW02009509 OWR50647.1 GAIX01014153 JAA78407.1 KQ971343 EFA04607.2 AK403273 BAM19673.1 AXCP01008274 GFDF01003939 JAV10145.1 GANO01001156 JAB58715.1 AJVK01059953 PYGN01000019 PSN57522.1 KYB27195.1 NEVH01006721 PNF37187.1 KK852543 KDR21662.1 GGFM01005994 MBW26745.1 GGFK01011277 MBW44598.1 ADMH02000253 ETN67235.1 AK417543 BAN20758.1 KQ459603 KPI92914.1 CH477434 GDUN01000301 EAT40979.1 JAN95618.1 GAPW01005676 GAPW01005675 JAC07923.1 GFDL01009980 JAV25065.1 DS232054 EDS33334.1 LJIG01016078 KRT81680.1 GEBQ01020357 JAT19620.1 GBBI01000515 JAC18197.1 GAHY01000866 JAA76644.1 AAAB01008984 EAA14788.4 APCN01003407 AXCN02000736 GFTR01002428 JAW13998.1 ATLV01025637 KE525396 KFB52499.1 JXUM01033516 KQ560991 KXJ80103.1 GL732664 EFX67993.1 KK854227 PTY13353.1 GDIP01094462 GDIQ01020652 JAM09253.1 JAN74085.1 LRGB01000930 KZS15100.1 IACF01001162 LAB66878.1 LNIX01000003 OXA57647.1 GBHO01033035 GBHO01022712 GBHO01022708 GBHO01022707 GBHO01022704 GBHO01022703 GBHO01019974 GBHO01019973 GBHO01019968 GBHO01011016 GBHO01011015 GBHO01011013 GBRD01001694 GBRD01001693 GDHC01013935 GDHC01009323 JAG10569.1 JAG20892.1 JAG20896.1 JAG20897.1 JAG20900.1 JAG20901.1 JAG23630.1 JAG23631.1 JAG23636.1 JAG32588.1 JAG32589.1 JAG32591.1 JAG64127.1 JAQ04694.1 JAQ09306.1 UFQS01000637 UFQT01000637 SSX05640.1 SSX25999.1 GDIQ01214870 JAK36855.1 APGK01046742 BT126797 KB741067 KB631751 AEE61759.1 ENN74410.1 ERL85793.1 GDIQ01040531 JAN54206.1 LJIJ01001181 ODM92633.1 DS235354 EEB15097.1 GDRN01053657 JAI66152.1 OUUW01000006 SPP81478.1 CH379059 EAL34193.2 CH479187 EDW39904.1 JRES01001119 KNC25393.1 GBYB01009746 JAG79513.1 CM000361 CM002910 EDX05209.1 KMY90498.1 CH902620 EDV31377.1 KPU73341.1 GDIQ01227321 GDIP01055477 JAK24404.1 JAM48238.1 CH480820 EDW54574.1 AE014134 KX531432 AGB93030.1 ANY27242.1 U28069 BT011529 GFDG01000806 JAV17993.1 CP012523 ALC39756.1 CH963852 EDW75650.1 CM000157 EDW89669.1 CH954177 EDV58099.1 EZ424090 ADD20366.1 GDHF01030694 JAI21620.1 KK122098 KFM81922.1 GAKP01010660 JAC48292.1 GGMR01000024 MBY12643.1 KQ414613 KOC68938.1 CH940649 KRF81350.1 CH933807 EDW11304.1 KRG02634.1 BT080175 ACO14599.1 ABLF02038238 KRG02633.1 GDIQ01060030 JAN34707.1 CAEY01001369 GFXV01002067 MBW13872.1 BT078302 BT121519 ACO12726.1 ADD38449.1 GDIP01013487 JAM90228.1 JH816867 EKC29744.1 AMQN01011676 KB309022 ELT95757.1
AGBW02009509 OWR50647.1 GAIX01014153 JAA78407.1 KQ971343 EFA04607.2 AK403273 BAM19673.1 AXCP01008274 GFDF01003939 JAV10145.1 GANO01001156 JAB58715.1 AJVK01059953 PYGN01000019 PSN57522.1 KYB27195.1 NEVH01006721 PNF37187.1 KK852543 KDR21662.1 GGFM01005994 MBW26745.1 GGFK01011277 MBW44598.1 ADMH02000253 ETN67235.1 AK417543 BAN20758.1 KQ459603 KPI92914.1 CH477434 GDUN01000301 EAT40979.1 JAN95618.1 GAPW01005676 GAPW01005675 JAC07923.1 GFDL01009980 JAV25065.1 DS232054 EDS33334.1 LJIG01016078 KRT81680.1 GEBQ01020357 JAT19620.1 GBBI01000515 JAC18197.1 GAHY01000866 JAA76644.1 AAAB01008984 EAA14788.4 APCN01003407 AXCN02000736 GFTR01002428 JAW13998.1 ATLV01025637 KE525396 KFB52499.1 JXUM01033516 KQ560991 KXJ80103.1 GL732664 EFX67993.1 KK854227 PTY13353.1 GDIP01094462 GDIQ01020652 JAM09253.1 JAN74085.1 LRGB01000930 KZS15100.1 IACF01001162 LAB66878.1 LNIX01000003 OXA57647.1 GBHO01033035 GBHO01022712 GBHO01022708 GBHO01022707 GBHO01022704 GBHO01022703 GBHO01019974 GBHO01019973 GBHO01019968 GBHO01011016 GBHO01011015 GBHO01011013 GBRD01001694 GBRD01001693 GDHC01013935 GDHC01009323 JAG10569.1 JAG20892.1 JAG20896.1 JAG20897.1 JAG20900.1 JAG20901.1 JAG23630.1 JAG23631.1 JAG23636.1 JAG32588.1 JAG32589.1 JAG32591.1 JAG64127.1 JAQ04694.1 JAQ09306.1 UFQS01000637 UFQT01000637 SSX05640.1 SSX25999.1 GDIQ01214870 JAK36855.1 APGK01046742 BT126797 KB741067 KB631751 AEE61759.1 ENN74410.1 ERL85793.1 GDIQ01040531 JAN54206.1 LJIJ01001181 ODM92633.1 DS235354 EEB15097.1 GDRN01053657 JAI66152.1 OUUW01000006 SPP81478.1 CH379059 EAL34193.2 CH479187 EDW39904.1 JRES01001119 KNC25393.1 GBYB01009746 JAG79513.1 CM000361 CM002910 EDX05209.1 KMY90498.1 CH902620 EDV31377.1 KPU73341.1 GDIQ01227321 GDIP01055477 JAK24404.1 JAM48238.1 CH480820 EDW54574.1 AE014134 KX531432 AGB93030.1 ANY27242.1 U28069 BT011529 GFDG01000806 JAV17993.1 CP012523 ALC39756.1 CH963852 EDW75650.1 CM000157 EDW89669.1 CH954177 EDV58099.1 EZ424090 ADD20366.1 GDHF01030694 JAI21620.1 KK122098 KFM81922.1 GAKP01010660 JAC48292.1 GGMR01000024 MBY12643.1 KQ414613 KOC68938.1 CH940649 KRF81350.1 CH933807 EDW11304.1 KRG02634.1 BT080175 ACO14599.1 ABLF02038238 KRG02633.1 GDIQ01060030 JAN34707.1 CAEY01001369 GFXV01002067 MBW13872.1 BT078302 BT121519 ACO12726.1 ADD38449.1 GDIP01013487 JAM90228.1 JH816867 EKC29744.1 AMQN01011676 KB309022 ELT95757.1
Proteomes
UP000037510
UP000283053
UP000007151
UP000007266
UP000075880
UP000092462
+ More
UP000245037 UP000235965 UP000027135 UP000000673 UP000069272 UP000053268 UP000008820 UP000002320 UP000075884 UP000075900 UP000075920 UP000075903 UP000076407 UP000007062 UP000075840 UP000075886 UP000030765 UP000069940 UP000249989 UP000000305 UP000192223 UP000076858 UP000076408 UP000075881 UP000075901 UP000075885 UP000075882 UP000002358 UP000198287 UP000019118 UP000030742 UP000094527 UP000009046 UP000268350 UP000001819 UP000008744 UP000037069 UP000095301 UP000095300 UP000000304 UP000192221 UP000007801 UP000001292 UP000000803 UP000092553 UP000007798 UP000002282 UP000008711 UP000054359 UP000053825 UP000008792 UP000009192 UP000091820 UP000007819 UP000015104 UP000005408 UP000014760
UP000245037 UP000235965 UP000027135 UP000000673 UP000069272 UP000053268 UP000008820 UP000002320 UP000075884 UP000075900 UP000075920 UP000075903 UP000076407 UP000007062 UP000075840 UP000075886 UP000030765 UP000069940 UP000249989 UP000000305 UP000192223 UP000076858 UP000076408 UP000075881 UP000075901 UP000075885 UP000075882 UP000002358 UP000198287 UP000019118 UP000030742 UP000094527 UP000009046 UP000268350 UP000001819 UP000008744 UP000037069 UP000095301 UP000095300 UP000000304 UP000192221 UP000007801 UP000001292 UP000000803 UP000092553 UP000007798 UP000002282 UP000008711 UP000054359 UP000053825 UP000008792 UP000009192 UP000091820 UP000007819 UP000015104 UP000005408 UP000014760
ProteinModelPortal
A0A0L7LM61
A0A3S2TPT5
A0A2H1UZK1
A0A212FA84
S4PS54
D6WLE9
+ More
I4DP33 A0A182JBA0 A0A1L8DV30 U5EM43 A0A1B0GPQ2 A0A2P8ZLZ4 A0A139WH69 A0A2J7R8M6 A0A067RD48 A0A2M3ZE16 A0A2M4AVF8 W5JVX0 A0A182F316 R4WDC1 A0A194PJL3 Q172M8 A0A023EE82 A0A1Q3FBV4 B0WRE4 A0A182NUP5 A0A0T6B2P7 A0A182RZB4 A0A182WGB7 A0A1B6L7H4 A0A023F9P2 R4G4D4 A0A182UXN3 A0A182WXL8 Q7PWG7 A0A182I7I2 A0A182QLC5 A0A224XVW3 A0A084WQK5 A0A182HCJ5 E9HJT3 A0A1W4WSK6 A0A2R7W0T9 A0A0P5VVU2 A0A164YC13 A0A2P2HYL8 A0A182Y0H4 A0A182KF06 A0A182T2V6 A0A182PVU2 A0A182LMJ8 K7J0R5 A0A226EKL1 A0A0A9YLW8 A0A336MAR6 A0A0P5IDQ6 J3JU84 A0A0P6G364 A0A1D2MI40 E0VNZ1 A0A0N7ZD36 A0A3B0KCW3 Q29NM5 B4GQN2 A0A0L0BZF8 A0A1I8MH61 A0A1I8Q4B7 A0A0C9RR36 B4Q701 A0A1W4VWF9 B3MJN8 A0A0P5YS29 B4I217 M9PDJ6 P49858 A0A1L8EHC4 A0A0M5J413 B4MU61 B4NXP4 B3N6D0 D3TRN8 A0A0K8U5G9 A0A087UX33 A0A034W1T8 A0A2S2N644 A0A0L7RDL0 A0A0Q9WAH1 B4KI22 P52159 C1BZZ6 A0A1A9WM33 J9JJ49 A0A0Q9X5X3 A0A0P6ENN8 T1K2W9 A0A2H8TJM0 C1BUM3 A0A0N8DPJ5 K1QF47 R7TQB5
I4DP33 A0A182JBA0 A0A1L8DV30 U5EM43 A0A1B0GPQ2 A0A2P8ZLZ4 A0A139WH69 A0A2J7R8M6 A0A067RD48 A0A2M3ZE16 A0A2M4AVF8 W5JVX0 A0A182F316 R4WDC1 A0A194PJL3 Q172M8 A0A023EE82 A0A1Q3FBV4 B0WRE4 A0A182NUP5 A0A0T6B2P7 A0A182RZB4 A0A182WGB7 A0A1B6L7H4 A0A023F9P2 R4G4D4 A0A182UXN3 A0A182WXL8 Q7PWG7 A0A182I7I2 A0A182QLC5 A0A224XVW3 A0A084WQK5 A0A182HCJ5 E9HJT3 A0A1W4WSK6 A0A2R7W0T9 A0A0P5VVU2 A0A164YC13 A0A2P2HYL8 A0A182Y0H4 A0A182KF06 A0A182T2V6 A0A182PVU2 A0A182LMJ8 K7J0R5 A0A226EKL1 A0A0A9YLW8 A0A336MAR6 A0A0P5IDQ6 J3JU84 A0A0P6G364 A0A1D2MI40 E0VNZ1 A0A0N7ZD36 A0A3B0KCW3 Q29NM5 B4GQN2 A0A0L0BZF8 A0A1I8MH61 A0A1I8Q4B7 A0A0C9RR36 B4Q701 A0A1W4VWF9 B3MJN8 A0A0P5YS29 B4I217 M9PDJ6 P49858 A0A1L8EHC4 A0A0M5J413 B4MU61 B4NXP4 B3N6D0 D3TRN8 A0A0K8U5G9 A0A087UX33 A0A034W1T8 A0A2S2N644 A0A0L7RDL0 A0A0Q9WAH1 B4KI22 P52159 C1BZZ6 A0A1A9WM33 J9JJ49 A0A0Q9X5X3 A0A0P6ENN8 T1K2W9 A0A2H8TJM0 C1BUM3 A0A0N8DPJ5 K1QF47 R7TQB5
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
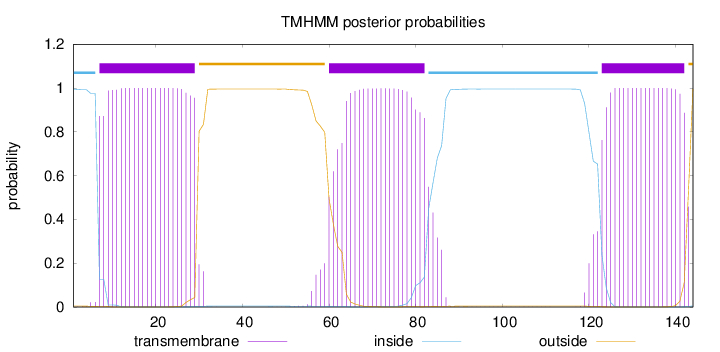
Length:
144
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.21236
Exp number, first 60 AAs:
24.14336
Total prob of N-in:
0.99561
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 59
TMhelix
60 - 82
inside
83 - 122
TMhelix
123 - 142
outside
143 - 144
Population Genetic Test Statistics
Pi
141.58356
Theta
145.633016
Tajima's D
-0.998225
CLR
0.438564
CSRT
0.142792860356982
Interpretation
Uncertain
