Gene
KWMTBOMO07079
Pre Gene Modal
BGIBMGA010433
Annotation
UDP-glycosyltransferase_UGT46A2_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 3.559
Sequence
CDS
ATGCGAGCTCACATCGCGTGCATCGTGCTGATCGCACACGCAGTCGCGTCCGCGAAAATATTAGGCCTTTTCCCGCACACAGGCAAAAGCCATCAAATGGTGTTCGACCCTTTATTGAGAAGATTGGCGGAGAAAGGCCACCATGTTACTGTAGTATCCTTTTTTCCCGTCAAAAATCCCCCAGCAAATTACACGAACATCAGCCTCGAGAGCTTAGCCAAATTAGGAGTTGAAACGATTGATTTGTCCTGGTATGAAAGCTCGAACTCCATACTCAAGATGACGGGACCGGAAAAAATATATGAAGAATTGACAGCGATGCAGCCGCTATCTTATATGGCCGTGGATGTGTGTTCCAAATTAGTTGGCTTCGAACCTTTGGCACGAACCTTGAGGCAGGATTACGATGTCATTTTAGTGGAGAATTTCAACAGTGACTGTATGCTGGGCCTGGCACATATATACGGTCAGAAAGCGCCAATTATAGCCTTATTATCAAGCTCGTTAATGGATTGGTCTCCGAGTCGTATTGGGGTATCAGATAATCCATCATACGTACCTATAGTCACTTCTACGTTTACTACCCCTATGTCGTTCGTGCAAAGATTGAAGAACACAGTTTTAAATATTTACTACAAGCTGTGGTTTCGTTACGCGATCCAATTGAAGGAGAAAGAAATAATTGAGAACCATTACGGTAGAAAGATCGTTGACCTCGAAGAATTAGCGAGAAATACGACATTAATGTTTGTTAACGTGCACCACAGTTTCAACGGTGTCAGACCTTTATTGCCCGGTATAGTAGAAGTGGGTGGCATGCATTTGGATCACAAAAGAAAGCCCATTCCAGAATTTTTCGAACGTTTCCTAAACGACTCTGAACACGGTGTAGTTTTATTTAGTTTTGGGTCCTTAATCAAGACATCTACGCTACCCAAGTACAAGGAGGATATTATAATGAAAACTCTTTCCCAACTGAAGCAGAGAGTTATCTGGAAATATGAAGATAGCGCTGAAGAAGGCACCCTTGTAGGAAACGTACTGAAAGTGAAGTGGATTCCTCAATATGATCTGTTACAGCATTCCAAAATAATTGCGTTCGTTGGGCACGGAGGACTTTTGGGCATGACCGAATCAATCTCAGCCGGCAAGCCCATGCTGGTGATACCGTTCTTCGGAGACCAGCACCTGAACGGCGCTCAAGCTGAAAAGATAGGGTTCGGTAAGGTGGTATCTTATGCTGATCTCTCGGAGAAGACGTTTCTGGATGGACTGCAAAGCGTGCTATCTCCTGAAATGCGATTGAGCGCACGAAGAGCGTCGAACATTTGGAGCGACCGACAAGCCGACCCGCTGGACACGGCCGTCTATTGGACAGAAAGGGTCATCCGTTGGGGACACCGAGCTCCCCTCCACTCGCCCGCGAGAGACTTGCCACTCCACCAGTATTTGTTGCTAGACGTTGCAGCAGCGATTTTAGTAGCAATTCTTGTGTTAATAGCGATTTTAAGATTGATTGTTGTATTGATAATCCGATTTTTCAGCGGTAGTGTCACTGCTAAAGAGAAATTACACTAA
Protein
MRAHIACIVLIAHAVASAKILGLFPHTGKSHQMVFDPLLRRLAEKGHHVTVVSFFPVKNPPANYTNISLESLAKLGVETIDLSWYESSNSILKMTGPEKIYEELTAMQPLSYMAVDVCSKLVGFEPLARTLRQDYDVILVENFNSDCMLGLAHIYGQKAPIIALLSSSLMDWSPSRIGVSDNPSYVPIVTSTFTTPMSFVQRLKNTVLNIYYKLWFRYAIQLKEKEIIENHYGRKIVDLEELARNTTLMFVNVHHSFNGVRPLLPGIVEVGGMHLDHKRKPIPEFFERFLNDSEHGVVLFSFGSLIKTSTLPKYKEDIIMKTLSQLKQRVIWKYEDSAEEGTLVGNVLKVKWIPQYDLLQHSKIIAFVGHGGLLGMTESISAGKPMLVIPFFGDQHLNGAQAEKIGFGKVVSYADLSEKTFLDGLQSVLSPEMRLSARRASNIWSDRQADPLDTAVYWTERVIRWGHRAPLHSPARDLPLHQYLLLDVAAAILVAILVLIAILRLIVVLIIRFFSGSVTAKEKLH
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPW1
G9LPW0
A0A2A4JGJ6
G9LPR6
A0A024AG07
A0A191T2K5
+ More
G9LPR7 A0A286MXP6 A0A2H4WB48 A0A194QN29 A0A212FA80 A0A194PHM3 A0A212FA68 A0A194QTP4 A0A194PP57 G9LPR8 A0A2H1VU07 A0A2A4JI28 A0A2J7R4W8 A0A2P8ZG78 A0A2J7R4W2 A0A067RAD1 A0A2J7R4S8 A0A1W4WYE4 D2A2V5 A0A2H4WB56 V5H2B2 A0A2J7R4W4 A0A1B6F5J1 A0A2R4FXJ3 A0A1W4WMC9 A0A182RBM8 A0A1L8D6F4 A0A182HV86 A0A1B6KXR8 A0A182YGV2 A0A2P8ZG71 A0A182WT93 A0A1S4GXF8 A0A1B6FW51 A0A2H4WB57 A0A084VP43 A0A0L7L3K3 W8AR85 A0A336KCF6 Q17HG0 A0A182V6Z3 A0A182GI26 A0A182W9Q0 A0A182UEM4 A0A1S4F2K0 A0A2J7R4T0 A0A182PJ64 A0A182NE44 A0A1W4WMF1 A0A182SC34 A0A182QSB1 A0A1I8N2C0 A0A1B6D7V3 A0A182IX39 A0A034V788 A0A1B6DCV6 A0A182F4M1 A0A2M3Z1Z1 T1PFI1 A0A0K8UM10 A0A2M3Z1Z8 W5J9T2 A0A0C9RNU1 A0A0K8UGE5 A0A1B6L8F2 A0A0K8VSF2 A0A151X0M2 A0A139WIV0 A0A0L0C9T9 A0A2S2NGA3 A0A191T1P6 Q7Q3K2 A0A2S2NJQ5 A0A0A1WW96 A0A1W7R8J2 A0A3S2LHF3 A0A2P8ZG70 B0WBP4 A0A1Q3FPW9 A0A139WIW7 F4WW48 A0A1Q3FPY1 A0A1I8NWF4 A0A1Y1LMI4
G9LPR7 A0A286MXP6 A0A2H4WB48 A0A194QN29 A0A212FA80 A0A194PHM3 A0A212FA68 A0A194QTP4 A0A194PP57 G9LPR8 A0A2H1VU07 A0A2A4JI28 A0A2J7R4W8 A0A2P8ZG78 A0A2J7R4W2 A0A067RAD1 A0A2J7R4S8 A0A1W4WYE4 D2A2V5 A0A2H4WB56 V5H2B2 A0A2J7R4W4 A0A1B6F5J1 A0A2R4FXJ3 A0A1W4WMC9 A0A182RBM8 A0A1L8D6F4 A0A182HV86 A0A1B6KXR8 A0A182YGV2 A0A2P8ZG71 A0A182WT93 A0A1S4GXF8 A0A1B6FW51 A0A2H4WB57 A0A084VP43 A0A0L7L3K3 W8AR85 A0A336KCF6 Q17HG0 A0A182V6Z3 A0A182GI26 A0A182W9Q0 A0A182UEM4 A0A1S4F2K0 A0A2J7R4T0 A0A182PJ64 A0A182NE44 A0A1W4WMF1 A0A182SC34 A0A182QSB1 A0A1I8N2C0 A0A1B6D7V3 A0A182IX39 A0A034V788 A0A1B6DCV6 A0A182F4M1 A0A2M3Z1Z1 T1PFI1 A0A0K8UM10 A0A2M3Z1Z8 W5J9T2 A0A0C9RNU1 A0A0K8UGE5 A0A1B6L8F2 A0A0K8VSF2 A0A151X0M2 A0A139WIV0 A0A0L0C9T9 A0A2S2NGA3 A0A191T1P6 Q7Q3K2 A0A2S2NJQ5 A0A0A1WW96 A0A1W7R8J2 A0A3S2LHF3 A0A2P8ZG70 B0WBP4 A0A1Q3FPW9 A0A139WIW7 F4WW48 A0A1Q3FPY1 A0A1I8NWF4 A0A1Y1LMI4
EC Number
2.4.1.17
Pubmed
EMBL
JQ070268
AEW43184.1
JQ070267
AEW43183.1
NWSH01001533
PCG70939.1
+ More
JQ070223 AEW43139.1 KF777119 AHY99690.1 KU680309 ANI22015.1 JQ070224 AEW43140.1 MF684364 ASX93998.1 KY202928 AUC64272.1 KQ461194 KPJ06917.1 AGBW02009509 OWR50646.1 KQ459603 KPI92916.1 OWR50645.1 KPJ06916.1 KPI92915.1 JQ070225 AEW43141.1 ODYU01004162 SOQ43734.1 NWSH01001496 PCG71080.1 NEVH01007402 PNF35870.1 PYGN01000068 PSN55501.1 PNF35872.1 KK852590 KDR20744.1 PNF35836.1 KQ971338 EFA02977.2 KY202929 AUC64273.1 GALX01001513 JAB66953.1 PNF35869.1 GECZ01024322 GECZ01008492 JAS45447.1 JAS61277.1 MF034860 AVT42226.1 GEYN01000096 JAV02033.1 APCN01002501 GEBQ01023933 JAT16044.1 PSN55491.1 AAAB01008964 GECZ01015337 JAS54432.1 KY202930 AUC64274.1 ATLV01014974 KE524999 KFB39737.1 JTDY01003254 KOB69869.1 GAMC01019493 GAMC01019492 GAMC01019491 JAB87063.1 UFQS01000219 UFQT01000219 SSX01542.1 SSX21922.1 CH477249 EAT46102.1 JXUM01065169 KQ562333 KXJ76125.1 PNF35837.1 AXCN02000871 GEDC01015537 JAS21761.1 GAKP01020965 JAC37987.1 GEDC01013767 JAS23531.1 GGFM01001770 MBW22521.1 KA647552 AFP62181.1 GDHF01024708 JAI27606.1 GGFM01001806 MBW22557.1 ADMH02001962 ETN60168.1 GBYB01008756 JAG78523.1 GDHF01026592 JAI25722.1 GEBQ01020086 JAT19891.1 GDHF01010498 JAI41816.1 KQ982611 KYQ53888.1 KYB27844.1 JRES01000714 KNC29017.1 GGMR01003614 MBY16233.1 KU680306 ANI22012.1 EAA12774.4 GGMR01004776 MBY17395.1 GBXI01011391 JAD02901.1 GEHC01000181 JAV47464.1 RSAL01000119 RVE46732.1 PSN55494.1 DS231880 EDS42579.1 GFDL01005396 JAV29649.1 KYB27856.1 GL888404 EGI61637.1 GFDL01005384 JAV29661.1 GEZM01054554 JAV73560.1
JQ070223 AEW43139.1 KF777119 AHY99690.1 KU680309 ANI22015.1 JQ070224 AEW43140.1 MF684364 ASX93998.1 KY202928 AUC64272.1 KQ461194 KPJ06917.1 AGBW02009509 OWR50646.1 KQ459603 KPI92916.1 OWR50645.1 KPJ06916.1 KPI92915.1 JQ070225 AEW43141.1 ODYU01004162 SOQ43734.1 NWSH01001496 PCG71080.1 NEVH01007402 PNF35870.1 PYGN01000068 PSN55501.1 PNF35872.1 KK852590 KDR20744.1 PNF35836.1 KQ971338 EFA02977.2 KY202929 AUC64273.1 GALX01001513 JAB66953.1 PNF35869.1 GECZ01024322 GECZ01008492 JAS45447.1 JAS61277.1 MF034860 AVT42226.1 GEYN01000096 JAV02033.1 APCN01002501 GEBQ01023933 JAT16044.1 PSN55491.1 AAAB01008964 GECZ01015337 JAS54432.1 KY202930 AUC64274.1 ATLV01014974 KE524999 KFB39737.1 JTDY01003254 KOB69869.1 GAMC01019493 GAMC01019492 GAMC01019491 JAB87063.1 UFQS01000219 UFQT01000219 SSX01542.1 SSX21922.1 CH477249 EAT46102.1 JXUM01065169 KQ562333 KXJ76125.1 PNF35837.1 AXCN02000871 GEDC01015537 JAS21761.1 GAKP01020965 JAC37987.1 GEDC01013767 JAS23531.1 GGFM01001770 MBW22521.1 KA647552 AFP62181.1 GDHF01024708 JAI27606.1 GGFM01001806 MBW22557.1 ADMH02001962 ETN60168.1 GBYB01008756 JAG78523.1 GDHF01026592 JAI25722.1 GEBQ01020086 JAT19891.1 GDHF01010498 JAI41816.1 KQ982611 KYQ53888.1 KYB27844.1 JRES01000714 KNC29017.1 GGMR01003614 MBY16233.1 KU680306 ANI22012.1 EAA12774.4 GGMR01004776 MBY17395.1 GBXI01011391 JAD02901.1 GEHC01000181 JAV47464.1 RSAL01000119 RVE46732.1 PSN55494.1 DS231880 EDS42579.1 GFDL01005396 JAV29649.1 KYB27856.1 GL888404 EGI61637.1 GFDL01005384 JAV29661.1 GEZM01054554 JAV73560.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000235965
UP000245037
+ More
UP000027135 UP000192223 UP000007266 UP000075900 UP000075840 UP000076408 UP000076407 UP000030765 UP000037510 UP000008820 UP000075903 UP000069940 UP000249989 UP000075920 UP000075902 UP000075885 UP000075884 UP000075901 UP000075886 UP000095301 UP000075880 UP000069272 UP000000673 UP000075809 UP000037069 UP000007062 UP000283053 UP000002320 UP000007755 UP000095300
UP000027135 UP000192223 UP000007266 UP000075900 UP000075840 UP000076408 UP000076407 UP000030765 UP000037510 UP000008820 UP000075903 UP000069940 UP000249989 UP000075920 UP000075902 UP000075885 UP000075884 UP000075901 UP000075886 UP000095301 UP000075880 UP000069272 UP000000673 UP000075809 UP000037069 UP000007062 UP000283053 UP000002320 UP000007755 UP000095300
Interpro
Gene 3D
ProteinModelPortal
G9LPW1
G9LPW0
A0A2A4JGJ6
G9LPR6
A0A024AG07
A0A191T2K5
+ More
G9LPR7 A0A286MXP6 A0A2H4WB48 A0A194QN29 A0A212FA80 A0A194PHM3 A0A212FA68 A0A194QTP4 A0A194PP57 G9LPR8 A0A2H1VU07 A0A2A4JI28 A0A2J7R4W8 A0A2P8ZG78 A0A2J7R4W2 A0A067RAD1 A0A2J7R4S8 A0A1W4WYE4 D2A2V5 A0A2H4WB56 V5H2B2 A0A2J7R4W4 A0A1B6F5J1 A0A2R4FXJ3 A0A1W4WMC9 A0A182RBM8 A0A1L8D6F4 A0A182HV86 A0A1B6KXR8 A0A182YGV2 A0A2P8ZG71 A0A182WT93 A0A1S4GXF8 A0A1B6FW51 A0A2H4WB57 A0A084VP43 A0A0L7L3K3 W8AR85 A0A336KCF6 Q17HG0 A0A182V6Z3 A0A182GI26 A0A182W9Q0 A0A182UEM4 A0A1S4F2K0 A0A2J7R4T0 A0A182PJ64 A0A182NE44 A0A1W4WMF1 A0A182SC34 A0A182QSB1 A0A1I8N2C0 A0A1B6D7V3 A0A182IX39 A0A034V788 A0A1B6DCV6 A0A182F4M1 A0A2M3Z1Z1 T1PFI1 A0A0K8UM10 A0A2M3Z1Z8 W5J9T2 A0A0C9RNU1 A0A0K8UGE5 A0A1B6L8F2 A0A0K8VSF2 A0A151X0M2 A0A139WIV0 A0A0L0C9T9 A0A2S2NGA3 A0A191T1P6 Q7Q3K2 A0A2S2NJQ5 A0A0A1WW96 A0A1W7R8J2 A0A3S2LHF3 A0A2P8ZG70 B0WBP4 A0A1Q3FPW9 A0A139WIW7 F4WW48 A0A1Q3FPY1 A0A1I8NWF4 A0A1Y1LMI4
G9LPR7 A0A286MXP6 A0A2H4WB48 A0A194QN29 A0A212FA80 A0A194PHM3 A0A212FA68 A0A194QTP4 A0A194PP57 G9LPR8 A0A2H1VU07 A0A2A4JI28 A0A2J7R4W8 A0A2P8ZG78 A0A2J7R4W2 A0A067RAD1 A0A2J7R4S8 A0A1W4WYE4 D2A2V5 A0A2H4WB56 V5H2B2 A0A2J7R4W4 A0A1B6F5J1 A0A2R4FXJ3 A0A1W4WMC9 A0A182RBM8 A0A1L8D6F4 A0A182HV86 A0A1B6KXR8 A0A182YGV2 A0A2P8ZG71 A0A182WT93 A0A1S4GXF8 A0A1B6FW51 A0A2H4WB57 A0A084VP43 A0A0L7L3K3 W8AR85 A0A336KCF6 Q17HG0 A0A182V6Z3 A0A182GI26 A0A182W9Q0 A0A182UEM4 A0A1S4F2K0 A0A2J7R4T0 A0A182PJ64 A0A182NE44 A0A1W4WMF1 A0A182SC34 A0A182QSB1 A0A1I8N2C0 A0A1B6D7V3 A0A182IX39 A0A034V788 A0A1B6DCV6 A0A182F4M1 A0A2M3Z1Z1 T1PFI1 A0A0K8UM10 A0A2M3Z1Z8 W5J9T2 A0A0C9RNU1 A0A0K8UGE5 A0A1B6L8F2 A0A0K8VSF2 A0A151X0M2 A0A139WIV0 A0A0L0C9T9 A0A2S2NGA3 A0A191T1P6 Q7Q3K2 A0A2S2NJQ5 A0A0A1WW96 A0A1W7R8J2 A0A3S2LHF3 A0A2P8ZG70 B0WBP4 A0A1Q3FPW9 A0A139WIW7 F4WW48 A0A1Q3FPY1 A0A1I8NWF4 A0A1Y1LMI4
PDB
2O6L
E-value=1.46841e-22,
Score=264
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 18,
Likelihood: 0.956728
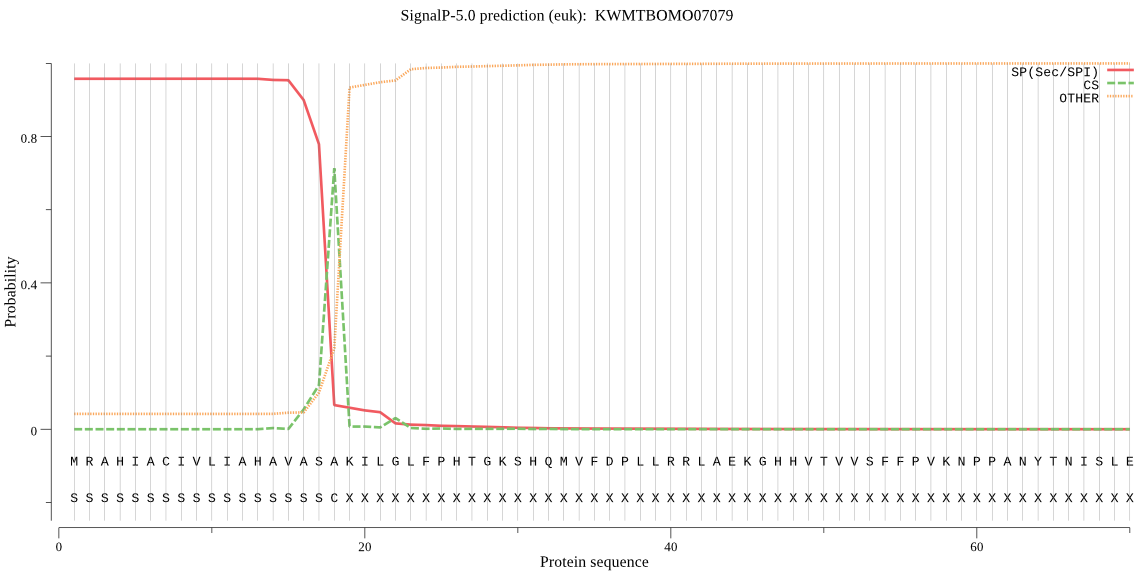
Length:
525
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.41431
Exp number, first 60 AAs:
15.18018
Total prob of N-in:
0.70626
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 488
TMhelix
489 - 511
inside
512 - 525

Population Genetic Test Statistics
Pi
146.745648
Theta
145.850112
Tajima's D
-0.780145
CLR
0.177594
CSRT
0.179391030448478
Interpretation
Uncertain
