Gene
KWMTBOMO07078
Pre Gene Modal
BGIBMGA010526
Annotation
PREDICTED:_KIF1-binding_protein_[Amyelois_transitella]
Full name
Protein KBP homolog
+ More
KIF1-binding protein
KIF1-binding protein
Location in the cell
Cytoplasmic Reliability : 2.037 Nuclear Reliability : 1.946
Sequence
CDS
ATGACTAGAGAAATGAGTGCTAAAGAGATACTTAATGATTTTAAAGAGAATTATATTAAATTCCGGAAGCTACTTGACGAAGACAGCAAGAATGATCCTGAAAATGCACCATATTTTTCTAAGTACAAAGCTAAAGAAATATTGAATAATATGTATGTTTTTTTAAAGACCTTAAAGACTGAAGGTTCCCCTGTCGATGATACCAAACTCGATGCTATGATCGGATATGTTTTGTTGAACATTGGTATAGTAGAAATGGATACAGAGGAACCTACTGCTAGTGAAAAGAGTCTCAATGAAGCTGTTGATCTCCTTCTGGAAAATGCTCTAAAACCCGAAATAGTTGTGACACTTATTAATGTCTACAACAATATTGGAATCTTATGGTCCAACAGAGATGACCCTGAAAAAGCAAAAGCTTCATTGGTTAAGGCAAAAACTTTGTATGAGGACTTTAAGTGCACTCTACAGATGCCATTACCTATAGAAGTCATTATAAATTATGTTGGGGAAACTTTAATAGATGATTTTATGTATTTGGAAAAATCTTATACTTTGACCTTGTATTATTTAGCTCAGGTGTTTGGTGTGTTAAAGGAGAACTTGAAATCAGCAATATATTGTCATGTAACATTAAGGAGACAGCTGGAATATAATGATTATGAACCTATAGACTGGGCATTGAATTCAGCTACATTGTCACAGTTCTTTGCTGAGCAAAATGGGTTTTACCAGTCAAGGAATCATTTAGCAGCTGCATCAACAATCCTTGATCAATATAAGCAACTGTTGGAGGCTGCAGTTGATCAAGATGAGGAATACTTAGCAAAACTAGAGACTTTTAAACATAGGTCTGCAGATGTTGCTAGGTGCTGGGCCAAATACTGTCTTTTATTGATGATAGCTTCAAAAACTAGGCTACTGGATGATGATGAAAAACTATCAGAGTCTATTACAGATATGTCAAACCTCAGCTTAGAAGACACTGAAAACATTTGTGAAGGTAATCTTAAGAAACTGACTTTTCCGAATCTCGATGTTACAAAATATGAAAGCAAAATAGGAGATAAGTACCTCCTGACTTATGAAGATGCTAGAGCAATTTTTTTAAGTTGCCAAAACTGGTTGAACCAAGCCAAGGAATATTATAAACTAGAAACGTTGGCTTCAGATTATATAGAACTTATTTTAGATAGCTCTCAGGCATACAATAATCTAGCATTTTTTGAGGATAATGATGATCGTAGGGCTAAAATGTTGAAGAGGCGAATTGATATGCTTGAAAACTTATTGAATGAAATAAATCCTACATATTATTTACAATACTGTAGACAGATTTGGTACGAACTCGGTGTTGTTTACACAGACATTTTGAACATAAAAATAGATAAATGGCCCAAGAGCGAAGCGACAGTTCAAGCAAGAAAAAAAATTGATTTGCTATGCAATAAAAGTATTGAAAGTTACACCAACTTTATTAATTCGATAAACGATAAAAATGGAAAAGTACCTGCAAAAATTGACCAGGAATTAATAAGACCTCTGATAAATGCTCTTGTTGCAGTCGGTAGGAACAGTATGAAACGCATGGCTGTAAATAAAATAAACCAATTGTGCCATGTTCAGAAAAGCCTTGATTCATACCAAACTGTTGTGGACATCTGTGATACACACGAAGAAGTTGCAGTGATGTTCACTGAAGAGTATAAGTTATGCCAAGAAATGTGTCAGATTTTACCTTTAAAAATCAAGAAGTTGCAAACCGAAATTGCTGGATAG
Protein
MTREMSAKEILNDFKENYIKFRKLLDEDSKNDPENAPYFSKYKAKEILNNMYVFLKTLKTEGSPVDDTKLDAMIGYVLLNIGIVEMDTEEPTASEKSLNEAVDLLLENALKPEIVVTLINVYNNIGILWSNRDDPEKAKASLVKAKTLYEDFKCTLQMPLPIEVIINYVGETLIDDFMYLEKSYTLTLYYLAQVFGVLKENLKSAIYCHVTLRRQLEYNDYEPIDWALNSATLSQFFAEQNGFYQSRNHLAAASTILDQYKQLLEAAVDQDEEYLAKLETFKHRSADVARCWAKYCLLLMIASKTRLLDDDEKLSESITDMSNLSLEDTENICEGNLKKLTFPNLDVTKYESKIGDKYLLTYEDARAIFLSCQNWLNQAKEYYKLETLASDYIELILDSSQAYNNLAFFEDNDDRRAKMLKRRIDMLENLLNEINPTYYLQYCRQIWYELGVVYTDILNIKIDKWPKSEATVQARKKIDLLCNKSIESYTNFINSINDKNGKVPAKIDQELIRPLINALVAVGRNSMKRMAVNKINQLCHVQKSLDSYQTVVDICDTHEEVAVMFTEEYKLCQEMCQILPLKIKKLQTEIAG
Summary
Description
Required for organization of axonal microtubules, and axonal outgrowth and maintenance during peripheral and central nervous system development.
Similarity
Belongs to the KIF1-binding protein family.
Keywords
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Reference proteome
Developmental protein
Differentiation
Neurogenesis
Feature
chain Protein KBP homolog
Uniprot
H9JLX4
A0A3S2M654
A0A2H1VDC0
A0A2A4K3B2
A0A194QPL2
A0A194PHX4
+ More
A0A0L7LF53 A0A212EH43 A0A0L7KLG9 A0A0T6B754 D2A380 A0A2J7R8M2 A0A2J7R8N7 N6TEK3 A0A1Y1K409 A0A1W4XG38 U4UHJ4 A0A2P8ZM20 A0A067RCU7 K7IVB4 A0A232FKC2 A0A1B6MDG1 E0W3X8 A0A1B6GF47 A0A310SEM4 A0A310SJT0 A0A088ANP6 A0A2A3EHZ6 A0A0M8ZUY1 A0A1B6C4E5 A0A1B0DB21 A0A182GK87 A0A1J1I787 A0A0L0C6X4 A0A0N8ES86 B0X4G8 A0A131XQC7 B7PKN4 A0A1I8NGH4 A0A1Q3FAB0 A0A1A9WKD6 V5IJ55 A0A0K8UAA2 A0A2M4CLL6 A0A182P952 A0A182MAY3 A0A2M4CM20 A0A034VX48 A0A1A9UPM7 E2C380 T1JN77 A0A2R5L4W6 A0A1A9ZP12 W8BG93 A0A182TI82 A0A182L5R0 Q5TVF2 A0A182V9F4 A0A182IE14 A0A182NEC2 A0A1S3IDL7 A0A182SPS0 A0A1B6HXP3 A0A0L7R3M2 A0A182Q2X4 A0A1L8DCH2 A0A182JD65 A0A182X7Q4 A0A182JYE6 Q29L23 W4XKH2 A0A182F4E0 A0A3R7NDC6 A0A2B4T1P8 A0A2R7WDK3 B4N0I1 H1UUN5 Q9VMX1 W5J2F8 A0A1W4UVK8 A0A084VPB1 A0A182RGH2 A0A0M4ESJ4 K1PRE5 B3MVL2 A0A224YM50 B3N4D1 A0A3B0K238 L7MJB0 A0A3M6TFW6 B4NW94 A0A0P4WFN4 A0A0J9QWU2 B4MDG5 B4Q358 A0A3Q2DZ74 R7V3X9 A0A060X7E2 Q0IIZ5 G3PUB0
A0A0L7LF53 A0A212EH43 A0A0L7KLG9 A0A0T6B754 D2A380 A0A2J7R8M2 A0A2J7R8N7 N6TEK3 A0A1Y1K409 A0A1W4XG38 U4UHJ4 A0A2P8ZM20 A0A067RCU7 K7IVB4 A0A232FKC2 A0A1B6MDG1 E0W3X8 A0A1B6GF47 A0A310SEM4 A0A310SJT0 A0A088ANP6 A0A2A3EHZ6 A0A0M8ZUY1 A0A1B6C4E5 A0A1B0DB21 A0A182GK87 A0A1J1I787 A0A0L0C6X4 A0A0N8ES86 B0X4G8 A0A131XQC7 B7PKN4 A0A1I8NGH4 A0A1Q3FAB0 A0A1A9WKD6 V5IJ55 A0A0K8UAA2 A0A2M4CLL6 A0A182P952 A0A182MAY3 A0A2M4CM20 A0A034VX48 A0A1A9UPM7 E2C380 T1JN77 A0A2R5L4W6 A0A1A9ZP12 W8BG93 A0A182TI82 A0A182L5R0 Q5TVF2 A0A182V9F4 A0A182IE14 A0A182NEC2 A0A1S3IDL7 A0A182SPS0 A0A1B6HXP3 A0A0L7R3M2 A0A182Q2X4 A0A1L8DCH2 A0A182JD65 A0A182X7Q4 A0A182JYE6 Q29L23 W4XKH2 A0A182F4E0 A0A3R7NDC6 A0A2B4T1P8 A0A2R7WDK3 B4N0I1 H1UUN5 Q9VMX1 W5J2F8 A0A1W4UVK8 A0A084VPB1 A0A182RGH2 A0A0M4ESJ4 K1PRE5 B3MVL2 A0A224YM50 B3N4D1 A0A3B0K238 L7MJB0 A0A3M6TFW6 B4NW94 A0A0P4WFN4 A0A0J9QWU2 B4MDG5 B4Q358 A0A3Q2DZ74 R7V3X9 A0A060X7E2 Q0IIZ5 G3PUB0
Pubmed
19121390
26354079
26227816
22118469
18362917
19820115
+ More
23537049 28004739 29403074 24845553 20075255 28648823 20566863 26483478 26108605 26999592 25315136 25765539 25348373 20798317 24495485 20966253 12364791 14747013 17210077 15632085 17994087 10731132 12537572 12537569 20920257 23761445 24438588 22992520 28797301 25576852 30382153 17550304 22936249 23254933 24755649
23537049 28004739 29403074 24845553 20075255 28648823 20566863 26483478 26108605 26999592 25315136 25765539 25348373 20798317 24495485 20966253 12364791 14747013 17210077 15632085 17994087 10731132 12537572 12537569 20920257 23761445 24438588 22992520 28797301 25576852 30382153 17550304 22936249 23254933 24755649
EMBL
BABH01005324
RSAL01000028
RVE51857.1
ODYU01001909
SOQ38776.1
NWSH01000232
+ More
PCG78160.1 KQ461194 KPJ06915.1 KQ459603 KPI92917.1 JTDY01001353 KOB74097.1 AGBW02014975 OWR40798.1 JTDY01009058 KOB64148.1 LJIG01009384 KRT83188.1 KQ971338 EFA01949.1 NEVH01006721 PNF37182.1 PNF37185.1 APGK01032042 KB740834 ENN78804.1 GEZM01098979 JAV53597.1 KB632166 ERL89335.1 PYGN01000019 PSN57546.1 KK852543 KDR21661.1 AAZX01013500 NNAY01000100 OXU30980.1 GEBQ01006004 JAT33973.1 DS235883 EEB20334.1 GECZ01008712 JAS61057.1 KQ785929 OAD47049.1 KQ759767 OAD62954.1 KZ288254 PBC30796.1 KQ435833 KOX71650.1 GEDC01028926 JAS08372.1 AJVK01000643 JXUM01013126 KQ560369 KXJ82756.1 CVRI01000043 CRK96120.1 JRES01000827 KNC28020.1 GDUN01000162 JAN95757.1 DS232340 EDS40321.1 GEFM01006547 JAP69249.1 ABJB010841772 DS734834 EEC07156.1 GFDL01010586 JAV24459.1 GANP01002027 JAB82441.1 GDHF01028798 JAI23516.1 GGFL01002054 MBW66232.1 AXCM01000251 GGFL01002053 MBW66231.1 GAKP01011934 GAKP01011933 JAC47019.1 GL452292 EFN77612.1 JH431868 GGLE01000361 MBY04487.1 GAMC01014244 JAB92311.1 AAAB01008839 EAL41373.2 APCN01004553 GECU01028277 JAS79429.1 KQ414663 KOC65351.1 AXCN02000864 GFDF01009913 JAV04171.1 CH379061 EAL33001.1 AAGJ04082373 AAGJ04082374 AAGJ04082375 QCYY01000588 ROT84145.1 LSMT01000002 PFX34708.1 KK854647 PTY17618.1 CH963920 EDW77594.1 BT133120 AEY64288.1 AE014134 BT011038 ADMH02002133 ETN58437.1 ATLV01014995 KE524999 KFB39805.1 CP012523 ALC40057.1 JH818888 EKC21399.1 CH902624 EDV33277.1 GFPF01003776 MAA14922.1 CH954177 EDV57801.1 OUUW01000006 SPP81950.1 GACK01000994 JAA64040.1 RCHS01003678 RMX40199.1 CM000157 EDW88411.1 GDRN01059426 JAI65497.1 CM002910 KMY88189.1 CH940661 EDW71226.1 CM000361 EDX03774.1 AMQN01005091 KB295123 ELU13558.1 FR905063 CDQ75563.1 BC121345
PCG78160.1 KQ461194 KPJ06915.1 KQ459603 KPI92917.1 JTDY01001353 KOB74097.1 AGBW02014975 OWR40798.1 JTDY01009058 KOB64148.1 LJIG01009384 KRT83188.1 KQ971338 EFA01949.1 NEVH01006721 PNF37182.1 PNF37185.1 APGK01032042 KB740834 ENN78804.1 GEZM01098979 JAV53597.1 KB632166 ERL89335.1 PYGN01000019 PSN57546.1 KK852543 KDR21661.1 AAZX01013500 NNAY01000100 OXU30980.1 GEBQ01006004 JAT33973.1 DS235883 EEB20334.1 GECZ01008712 JAS61057.1 KQ785929 OAD47049.1 KQ759767 OAD62954.1 KZ288254 PBC30796.1 KQ435833 KOX71650.1 GEDC01028926 JAS08372.1 AJVK01000643 JXUM01013126 KQ560369 KXJ82756.1 CVRI01000043 CRK96120.1 JRES01000827 KNC28020.1 GDUN01000162 JAN95757.1 DS232340 EDS40321.1 GEFM01006547 JAP69249.1 ABJB010841772 DS734834 EEC07156.1 GFDL01010586 JAV24459.1 GANP01002027 JAB82441.1 GDHF01028798 JAI23516.1 GGFL01002054 MBW66232.1 AXCM01000251 GGFL01002053 MBW66231.1 GAKP01011934 GAKP01011933 JAC47019.1 GL452292 EFN77612.1 JH431868 GGLE01000361 MBY04487.1 GAMC01014244 JAB92311.1 AAAB01008839 EAL41373.2 APCN01004553 GECU01028277 JAS79429.1 KQ414663 KOC65351.1 AXCN02000864 GFDF01009913 JAV04171.1 CH379061 EAL33001.1 AAGJ04082373 AAGJ04082374 AAGJ04082375 QCYY01000588 ROT84145.1 LSMT01000002 PFX34708.1 KK854647 PTY17618.1 CH963920 EDW77594.1 BT133120 AEY64288.1 AE014134 BT011038 ADMH02002133 ETN58437.1 ATLV01014995 KE524999 KFB39805.1 CP012523 ALC40057.1 JH818888 EKC21399.1 CH902624 EDV33277.1 GFPF01003776 MAA14922.1 CH954177 EDV57801.1 OUUW01000006 SPP81950.1 GACK01000994 JAA64040.1 RCHS01003678 RMX40199.1 CM000157 EDW88411.1 GDRN01059426 JAI65497.1 CM002910 KMY88189.1 CH940661 EDW71226.1 CM000361 EDX03774.1 AMQN01005091 KB295123 ELU13558.1 FR905063 CDQ75563.1 BC121345
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000037510
+ More
UP000007151 UP000007266 UP000235965 UP000019118 UP000192223 UP000030742 UP000245037 UP000027135 UP000002358 UP000215335 UP000009046 UP000005203 UP000242457 UP000053105 UP000092462 UP000069940 UP000249989 UP000183832 UP000037069 UP000002320 UP000001555 UP000095301 UP000091820 UP000075885 UP000075883 UP000078200 UP000008237 UP000092445 UP000075902 UP000075882 UP000007062 UP000075903 UP000075840 UP000075884 UP000085678 UP000075901 UP000053825 UP000075886 UP000075880 UP000076407 UP000075881 UP000001819 UP000007110 UP000069272 UP000283509 UP000225706 UP000007798 UP000000803 UP000000673 UP000192221 UP000030765 UP000075900 UP000092553 UP000005408 UP000007801 UP000008711 UP000268350 UP000275408 UP000002282 UP000008792 UP000000304 UP000265020 UP000014760 UP000193380 UP000008143 UP000007635
UP000007151 UP000007266 UP000235965 UP000019118 UP000192223 UP000030742 UP000245037 UP000027135 UP000002358 UP000215335 UP000009046 UP000005203 UP000242457 UP000053105 UP000092462 UP000069940 UP000249989 UP000183832 UP000037069 UP000002320 UP000001555 UP000095301 UP000091820 UP000075885 UP000075883 UP000078200 UP000008237 UP000092445 UP000075902 UP000075882 UP000007062 UP000075903 UP000075840 UP000075884 UP000085678 UP000075901 UP000053825 UP000075886 UP000075880 UP000076407 UP000075881 UP000001819 UP000007110 UP000069272 UP000283509 UP000225706 UP000007798 UP000000803 UP000000673 UP000192221 UP000030765 UP000075900 UP000092553 UP000005408 UP000007801 UP000008711 UP000268350 UP000275408 UP000002282 UP000008792 UP000000304 UP000265020 UP000014760 UP000193380 UP000008143 UP000007635
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JLX4
A0A3S2M654
A0A2H1VDC0
A0A2A4K3B2
A0A194QPL2
A0A194PHX4
+ More
A0A0L7LF53 A0A212EH43 A0A0L7KLG9 A0A0T6B754 D2A380 A0A2J7R8M2 A0A2J7R8N7 N6TEK3 A0A1Y1K409 A0A1W4XG38 U4UHJ4 A0A2P8ZM20 A0A067RCU7 K7IVB4 A0A232FKC2 A0A1B6MDG1 E0W3X8 A0A1B6GF47 A0A310SEM4 A0A310SJT0 A0A088ANP6 A0A2A3EHZ6 A0A0M8ZUY1 A0A1B6C4E5 A0A1B0DB21 A0A182GK87 A0A1J1I787 A0A0L0C6X4 A0A0N8ES86 B0X4G8 A0A131XQC7 B7PKN4 A0A1I8NGH4 A0A1Q3FAB0 A0A1A9WKD6 V5IJ55 A0A0K8UAA2 A0A2M4CLL6 A0A182P952 A0A182MAY3 A0A2M4CM20 A0A034VX48 A0A1A9UPM7 E2C380 T1JN77 A0A2R5L4W6 A0A1A9ZP12 W8BG93 A0A182TI82 A0A182L5R0 Q5TVF2 A0A182V9F4 A0A182IE14 A0A182NEC2 A0A1S3IDL7 A0A182SPS0 A0A1B6HXP3 A0A0L7R3M2 A0A182Q2X4 A0A1L8DCH2 A0A182JD65 A0A182X7Q4 A0A182JYE6 Q29L23 W4XKH2 A0A182F4E0 A0A3R7NDC6 A0A2B4T1P8 A0A2R7WDK3 B4N0I1 H1UUN5 Q9VMX1 W5J2F8 A0A1W4UVK8 A0A084VPB1 A0A182RGH2 A0A0M4ESJ4 K1PRE5 B3MVL2 A0A224YM50 B3N4D1 A0A3B0K238 L7MJB0 A0A3M6TFW6 B4NW94 A0A0P4WFN4 A0A0J9QWU2 B4MDG5 B4Q358 A0A3Q2DZ74 R7V3X9 A0A060X7E2 Q0IIZ5 G3PUB0
A0A0L7LF53 A0A212EH43 A0A0L7KLG9 A0A0T6B754 D2A380 A0A2J7R8M2 A0A2J7R8N7 N6TEK3 A0A1Y1K409 A0A1W4XG38 U4UHJ4 A0A2P8ZM20 A0A067RCU7 K7IVB4 A0A232FKC2 A0A1B6MDG1 E0W3X8 A0A1B6GF47 A0A310SEM4 A0A310SJT0 A0A088ANP6 A0A2A3EHZ6 A0A0M8ZUY1 A0A1B6C4E5 A0A1B0DB21 A0A182GK87 A0A1J1I787 A0A0L0C6X4 A0A0N8ES86 B0X4G8 A0A131XQC7 B7PKN4 A0A1I8NGH4 A0A1Q3FAB0 A0A1A9WKD6 V5IJ55 A0A0K8UAA2 A0A2M4CLL6 A0A182P952 A0A182MAY3 A0A2M4CM20 A0A034VX48 A0A1A9UPM7 E2C380 T1JN77 A0A2R5L4W6 A0A1A9ZP12 W8BG93 A0A182TI82 A0A182L5R0 Q5TVF2 A0A182V9F4 A0A182IE14 A0A182NEC2 A0A1S3IDL7 A0A182SPS0 A0A1B6HXP3 A0A0L7R3M2 A0A182Q2X4 A0A1L8DCH2 A0A182JD65 A0A182X7Q4 A0A182JYE6 Q29L23 W4XKH2 A0A182F4E0 A0A3R7NDC6 A0A2B4T1P8 A0A2R7WDK3 B4N0I1 H1UUN5 Q9VMX1 W5J2F8 A0A1W4UVK8 A0A084VPB1 A0A182RGH2 A0A0M4ESJ4 K1PRE5 B3MVL2 A0A224YM50 B3N4D1 A0A3B0K238 L7MJB0 A0A3M6TFW6 B4NW94 A0A0P4WFN4 A0A0J9QWU2 B4MDG5 B4Q358 A0A3Q2DZ74 R7V3X9 A0A060X7E2 Q0IIZ5 G3PUB0
Ontologies
PANTHER
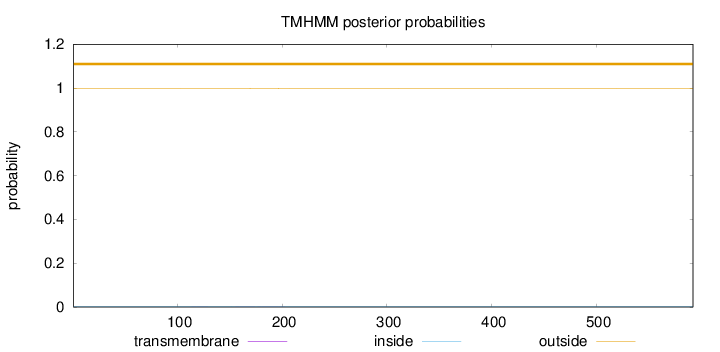
Topology
Subcellular location
Length:
592
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01999
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.00282
outside
1 - 592
Population Genetic Test Statistics
Pi
192.356296
Theta
196.06351
Tajima's D
-0.125376
CLR
0
CSRT
0.333833308334583
Interpretation
Uncertain
