Pre Gene Modal
BGIBMGA010525
Annotation
putative_mesoderm_induction_early_response_1_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.112
Sequence
CDS
ATGTCAGATTGCGCGCTGGTTACCAATGTAAGCGAACACGATGCGAGCATGGATGTGGGGAGCGACAAGTCTCTATTCGAGCCTACTGTTGATATGATGGTCAATGATTTCGATGACGAGAGAACTTTAGACGAAGAAGAAGCTTTAGCTGCAGGCGAGCAACAGGACCCTAAGGCCGAGCTCAATAGCCTTCAGAAGGAGGGAGATATGCCTTTAGAGGAACTCCTGGCTCTGTACGGTTACGACAGAAACATGGAAAAGCCAATTCCTGACCAAGTTCCAGAAGTAGTTCCTGAGGAAACTGAGAGAACGGAGAGGGCTGAGAGGACTGAGAGAAATGAATCTGCCCTTTCCCAATTATACAATGAGAATGCAAACCCTGAAGCTACTAGATGCCTTCGGTCTGGCTCAAGGCCTCCCTCTGAGGAGGAAGATGATTATGATTACAGTCCAGATGAGGAAGACTGGAAAAAAACTATAATGGTTGGTAGTGACTATCAAGCTGCTATACCTGAGGGTCTATGCAGCTATGATGATGCATTACCTTATGAAAATGAAGATAAATTACTATGGAATCCTTCTATTCTTGATGAAAGAATCACTGAAGAGTACATGCGAAAAGTGTGTGCTCTGAATTTAGGCACCGGGATTGAATTAATTCCAAAAGGAAAACAGCTTCGTGATGATGAAGAGGCCCTGTACTTATTGCAACAATGTGGCCACAATGCAGAGGAAGCCTTGAGACGGAAACGCATTTCTGCACAAACTCCACCTCACGCAAGTCTTTGGTCAGAAGAAGAATGCCGAAATTTTGAAAATGGCCTAAAAGCACACGGAAAAGACTTTCACTTGATAAGGCAGAACAAAGTACGAACTCGGTCTGTCGGAGAACTCGTCCAATTCTACTATATTTGGAAAAAAACTGAAAGGCATGATATATTTGCAAATAAGGCTAGATTGGAGAAGAAAAAGTATACTTTGCATCCTGGTCACACTGATTACATGGATAGGTTCTTAGAGGAGCAAGAGGCAAGTGGAGGAAGTGTGGTGCGACCTGCATCTCCATCGCCCATGATGGACTATGCAACCTCTCCTGTCACTCAACCCGACCCTCTTGCTCTTGGTGAGAAAGAAGTATTTTCACAGTTAAATACACATAACACACCCCCGAGGGCCATGTCGACAGAAGAACCAGAAGCAGACACTGGACCGTAA
Protein
MSDCALVTNVSEHDASMDVGSDKSLFEPTVDMMVNDFDDERTLDEEEALAAGEQQDPKAELNSLQKEGDMPLEELLALYGYDRNMEKPIPDQVPEVVPEETERTERAERTERNESALSQLYNENANPEATRCLRSGSRPPSEEEDDYDYSPDEEDWKKTIMVGSDYQAAIPEGLCSYDDALPYENEDKLLWNPSILDERITEEYMRKVCALNLGTGIELIPKGKQLRDDEEALYLLQQCGHNAEEALRRKRISAQTPPHASLWSEEECRNFENGLKAHGKDFHLIRQNKVRTRSVGELVQFYYIWKKTERHDIFANKARLEKKKYTLHPGHTDYMDRFLEEQEASGGSVVRPASPSPMMDYATSPVTQPDPLALGEKEVFSQLNTHNTPPRAMSTEEPEADTGP
Summary
Uniprot
H9JLX3
A0A1E1W2R7
A0A2A4K1L6
A0A2H1VD72
A0A194PIP1
A0A194QNI1
+ More
A0A212EH31 A0A0L7LTU4 S4P5L6 A0A067QU08 A0A2P8YMV7 A0A2J7PM97 A0A2J7PMC1 A0A1Y1LH32 A0A023F219 T1HAI2 D7EL50 A0A0A9YQZ7 A0A0A9Y7H3 A0A139WPQ0 A0A210QJB9 A0A224XSE5 A0A2R7W0Y7 E0VPL9 R7VGZ9 A0A0T6AY00 A0A0L8FRT5 V3ZVR1 A0A0L8FRW7 A0A146LT57 A0A0B6YCI5 A0A0B6YB39
A0A212EH31 A0A0L7LTU4 S4P5L6 A0A067QU08 A0A2P8YMV7 A0A2J7PM97 A0A2J7PMC1 A0A1Y1LH32 A0A023F219 T1HAI2 D7EL50 A0A0A9YQZ7 A0A0A9Y7H3 A0A139WPQ0 A0A210QJB9 A0A224XSE5 A0A2R7W0Y7 E0VPL9 R7VGZ9 A0A0T6AY00 A0A0L8FRT5 V3ZVR1 A0A0L8FRW7 A0A146LT57 A0A0B6YCI5 A0A0B6YB39
Pubmed
EMBL
BABH01005324
GDQN01009810
JAT81244.1
NWSH01000232
PCG78161.1
ODYU01001909
+ More
SOQ38777.1 KQ459603 KPI92918.1 KQ461194 KPJ06914.1 AGBW02014975 OWR40797.1 JTDY01000095 KOB78903.1 GAIX01008532 JAA84028.1 KK852941 KDR13525.1 PYGN01000482 PSN45590.1 NEVH01024418 PNF17461.1 PNF17459.1 GEZM01060090 GEZM01060088 GEZM01060085 GEZM01060083 JAV71275.1 GBBI01003100 JAC15612.1 ACPB03011331 DS497806 EFA11881.1 GBHO01010081 GBHO01010079 JAG33523.1 JAG33525.1 GBHO01014572 JAG29032.1 KQ973148 KYB29912.1 NEDP02003363 OWF48842.1 GFTR01005527 JAW10899.1 KK854175 PTY12600.1 DS235366 EEB15325.1 AMQN01016493 KB292122 ELU18108.1 LJIG01022642 KRT79479.1 KQ427059 KOF67431.1 KB203019 ESO86695.1 KOF67429.1 GDHC01009009 JAQ09620.1 HACG01006671 CEK53536.1 HACG01006672 CEK53537.1
SOQ38777.1 KQ459603 KPI92918.1 KQ461194 KPJ06914.1 AGBW02014975 OWR40797.1 JTDY01000095 KOB78903.1 GAIX01008532 JAA84028.1 KK852941 KDR13525.1 PYGN01000482 PSN45590.1 NEVH01024418 PNF17461.1 PNF17459.1 GEZM01060090 GEZM01060088 GEZM01060085 GEZM01060083 JAV71275.1 GBBI01003100 JAC15612.1 ACPB03011331 DS497806 EFA11881.1 GBHO01010081 GBHO01010079 JAG33523.1 JAG33525.1 GBHO01014572 JAG29032.1 KQ973148 KYB29912.1 NEDP02003363 OWF48842.1 GFTR01005527 JAW10899.1 KK854175 PTY12600.1 DS235366 EEB15325.1 AMQN01016493 KB292122 ELU18108.1 LJIG01022642 KRT79479.1 KQ427059 KOF67431.1 KB203019 ESO86695.1 KOF67429.1 GDHC01009009 JAQ09620.1 HACG01006671 CEK53536.1 HACG01006672 CEK53537.1
Proteomes
PRIDE
Pfam
PF01448 ELM2
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
H9JLX3
A0A1E1W2R7
A0A2A4K1L6
A0A2H1VD72
A0A194PIP1
A0A194QNI1
+ More
A0A212EH31 A0A0L7LTU4 S4P5L6 A0A067QU08 A0A2P8YMV7 A0A2J7PM97 A0A2J7PMC1 A0A1Y1LH32 A0A023F219 T1HAI2 D7EL50 A0A0A9YQZ7 A0A0A9Y7H3 A0A139WPQ0 A0A210QJB9 A0A224XSE5 A0A2R7W0Y7 E0VPL9 R7VGZ9 A0A0T6AY00 A0A0L8FRT5 V3ZVR1 A0A0L8FRW7 A0A146LT57 A0A0B6YCI5 A0A0B6YB39
A0A212EH31 A0A0L7LTU4 S4P5L6 A0A067QU08 A0A2P8YMV7 A0A2J7PM97 A0A2J7PMC1 A0A1Y1LH32 A0A023F219 T1HAI2 D7EL50 A0A0A9YQZ7 A0A0A9Y7H3 A0A139WPQ0 A0A210QJB9 A0A224XSE5 A0A2R7W0Y7 E0VPL9 R7VGZ9 A0A0T6AY00 A0A0L8FRT5 V3ZVR1 A0A0L8FRW7 A0A146LT57 A0A0B6YCI5 A0A0B6YB39
PDB
4CZZ
E-value=6.55806e-08,
Score=136
Ontologies
GO
PANTHER
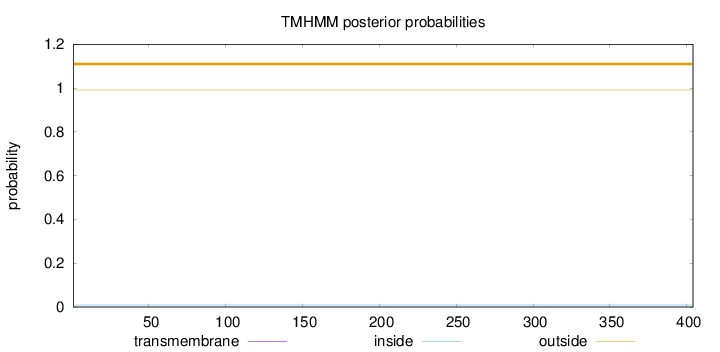
Topology
Subcellular location
Nucleus
Length:
404
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00903
outside
1 - 404
Population Genetic Test Statistics
Pi
149.978686
Theta
161.289464
Tajima's D
-1.138661
CLR
259.72737
CSRT
0.114494275286236
Interpretation
Uncertain
