Gene
KWMTBOMO07075
Pre Gene Modal
BGIBMGA010524
Annotation
PREDICTED:_engulfment_and_cell_motility_protein_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.774 Nuclear Reliability : 2.429
Sequence
CDS
ATGTCTACAAAACCGATAAAAATGCCGTCGGCAACAAAAGAAAAGGATTCTACGAAACTTAAGATTGCTGTGGAAATGTTAAATGCTCCAAATAAAGTACCACAGCTCATCGAGTTAGATCAAAGCCAGCCTTTATCGAGCATAATCCAGCTTCTTTGTAAAACGTGGGGACTTCCGGACCCCGTGAACTACGCTTTGCAGTTCTCTGAGAGCAATAACCAAAACTACATTACCGAGAAAAACAGGAATGAGATAAAGAACGGTTCAGTGTTACGCCTAGAGTTATCGCCTGCGAAAACGGTGCAAGACATTTTGGCCAAGATCAATGGCGTAGAAGCAGATCAAATCGCCGCCTTATCCAAATTATCTGTCCTTAGTAGCGACCTAACATTTGCATTAGAGTTCATTAACAAAAAAGGTCTGACACTCATCATCAATGACATCGAAAATGGAAAATTCAAAGGAAAATCGCTAAAATACGCCTTAGCAACTTTTGTTGAACTCATGGACCACAGTGTAATATTCTGGGATATACTAGAAAATCAATTCATAAACAAAATCGCCAGCTTTGTGAGTAACCAGGCCATTCCGCAAGACCCCAAAGTAATTCAGTGTTGCTTGTCCATTCTAGAAAACATTGTACTGAACAGCTCTAAGAACTCTCAAGTGGAAAAAGAAATCACAATTCCAAGTTTGATATCACATTTACAGAATTCACAACAGGACAGTTTAATTCAGCAGAATGCTATTGCTCTCATAAATGCTATATTTTGCAAAGCCAATATGACAAAGAAAAAAACTATTGCAGCAACTTTGTCATCTAAACAGGTACGTAATGTCATTTATGAGAACCTCATTGGGACTGGTGAGTTCCCTAAGAACAATACTAAAGAAGCATTAGGGACTGAATTGGCTCACCAGCTCTATGTACTTCAAACATTGATGCTGGGTTTGCTGGAGCCACAAATGAGACAAAAGGCTGAGTCACAGGAACAAGAGTCACAAGAAAAAATAAAGGAATTGAGGAAAATTGCTTTTGAAGGTGACAACAATCCAAATATTGAAATAACAATGAGAAGACCTGTTGGTTCTTATTCAAAAGATTTCAAAAAACTTGGCTTTAAATGTGAAATAGATCCTATTAAAGATTTCAACGAGACTCCCCCAGGAGTACTGGCATTAGACTGTATGTTGTATTTTGCCAGAAATTACCCAGAAGATTATACTAAAATAGTTCTTGAAAATAGCTGCCGTGCAGACGAACATGAGTGTCCCTTTGGGAAGACAAGCGTGGAGCTTGTAAAGCAACTGTGTGACATCCTACATGTAGGTGAACCACCAAGCGAACAAGGCCAGACATACCATCCTCTCTTCTTCACACATGATCATCCATTTGAAGAACTATTCTGCATTTGTATAGTTCTACTTAATAAGACATGGAAGGAAATGAGAGCGACATCTGAAGACTTCATAAAGGTATCGAGTGTCGTAAGAGAACAGATTAGCAGAGCGCTCTTAGCATCACCTAAAAACTTTGATAAGTTCCGGCAAAAAATTAACCAGCTGACATATGCCGAAATAACACAAATATGGCAGCAAGAAAGAACGAACAGAGAAGTCTGGGAGTCTCATGCGCGACCTATTGTAGAGTTAAAAGAAAAAATTACACCAGAAATTGTAGACTTGATTCAGCAACAGAGATTGGGTGTACTTGTTGGTGGAACAAGGTTTAAAAAGTTTTTTTCTAGAAGTCAAAGAATAAAAGATAAGTTCTGGTTTGTTCGTCTCTCACCTAACTATAAAGTGTTGCACTATGGTGATTGTGATGAAAAAAGCACTCCGAGCTTAGAAGAACTCGGAAACAAGCTACCTGTTGCTGAAATCAAATGTGTTGTTGTCGGTAAAGAGTGTCCACACATGAAGGACTTGAGAGGAAGAAAAATAACACCACATCTGGCTTTTTCATTTATTTTGAAATCTGCTGAAGTTACTTCTTTAGATTTTGTGGCTCCTGATGAACAGATCTTCGACTACTGGACTGATGGTATCAATGCACTCTTAAAGGAAAAGATGACCAGTAAATCATTTGAAAACGATTTGCAAACTCTACTGTCCATGGATATAAAGGTTCGGTTACTTGATGCGGAAGGCATTGACATTCCACAAGATCCTCCACAAATTCCTGATGAACCTGAAGATTATGATTTTTATTATGAGAATAATTAG
Protein
MSTKPIKMPSATKEKDSTKLKIAVEMLNAPNKVPQLIELDQSQPLSSIIQLLCKTWGLPDPVNYALQFSESNNQNYITEKNRNEIKNGSVLRLELSPAKTVQDILAKINGVEADQIAALSKLSVLSSDLTFALEFINKKGLTLIINDIENGKFKGKSLKYALATFVELMDHSVIFWDILENQFINKIASFVSNQAIPQDPKVIQCCLSILENIVLNSSKNSQVEKEITIPSLISHLQNSQQDSLIQQNAIALINAIFCKANMTKKKTIAATLSSKQVRNVIYENLIGTGEFPKNNTKEALGTELAHQLYVLQTLMLGLLEPQMRQKAESQEQESQEKIKELRKIAFEGDNNPNIEITMRRPVGSYSKDFKKLGFKCEIDPIKDFNETPPGVLALDCMLYFARNYPEDYTKIVLENSCRADEHECPFGKTSVELVKQLCDILHVGEPPSEQGQTYHPLFFTHDHPFEELFCICIVLLNKTWKEMRATSEDFIKVSSVVREQISRALLASPKNFDKFRQKINQLTYAEITQIWQQERTNREVWESHARPIVELKEKITPEIVDLIQQQRLGVLVGGTRFKKFFSRSQRIKDKFWFVRLSPNYKVLHYGDCDEKSTPSLEELGNKLPVAEIKCVVVGKECPHMKDLRGRKITPHLAFSFILKSAEVTSLDFVAPDEQIFDYWTDGINALLKEKMTSKSFENDLQTLLSMDIKVRLLDAEGIDIPQDPPQIPDEPEDYDFYYENN
Summary
Uniprot
H9JLX2
A0A2A4K309
A0A2H1VQ83
A0A1E1W7S9
A0A194QN25
A0A194PP61
+ More
A0A0L7LUK9 A0A212EH40 A0A2J7R8I2 A0A067QYM9 A0A0J7KR76 A0A158NYE5 A0A026VTW0 E2ARV2 A0A2A3EFB4 A0A195F6A6 A0A087ZYU3 A0A0M8ZZY0 A0A151IEZ9 F4WT65 A0A151J547 A0A310SUR3 E2CAD6 E0VNX9 A0A1B6F7R2 E9JD71 A0A1B6DW40 A0A232F3Y7 K7J0J3 A0A0C9RD96 A0A0L7RGV4 A0A154PQ91 T1I4G8 J9JPM5 A0A023F487 A0A1Y1L2K6 A0A2S2NQB3 A0A2S2R4K8 D6WDN2 A0A0C9PU86 A0A1W4X9P2 A0A224X8S2 A0A0A9WC65 A0A2P6L738 A0A182WHB2 A0A1B0CX19 A0A182M032 A0A182RG46 A0A182Q9P6 A0A084VY71 A0A182I5B6 Q7PVN1 A0A182VGL6 A0A182UAQ9 A0A182KU93 A0A182XKS9 A0A3L8DUU7 A0A182KC16 A0A2L2YET9 A0A0C9QW66 A0A182PAN0 A0A182Y9X1 U5ELH0 A0A182NF89 A0A182J4L7 T1J981 A0A1E1X548 A0A2M4AN54 A0A131YNT2 A0A224YY59 A0A1E1XMB8 W5JP59 B7Q7M8 A0A1B0DC22 L7M9U3 B0WB15 A0A182F121 L7LVG7 Q17LW5 A0A0P5KTU8 A0A0N8C9N4 A0A1W4UYR8 A0A0P5YWD9 A0A0P6IA13 A0A1I8NBP5 A0A0P5KII8 B4KI03 T1P995 A0A1A9XEP8 B3N3X1 E9H9Z7 B4P1Y7 A0A1A9WKK9 W8B5U8 A0A1B0FEC7 A0A1A9V886 A0A3B0K2J4 A0A034VW87 B4IE65 B4Q3Q3
A0A0L7LUK9 A0A212EH40 A0A2J7R8I2 A0A067QYM9 A0A0J7KR76 A0A158NYE5 A0A026VTW0 E2ARV2 A0A2A3EFB4 A0A195F6A6 A0A087ZYU3 A0A0M8ZZY0 A0A151IEZ9 F4WT65 A0A151J547 A0A310SUR3 E2CAD6 E0VNX9 A0A1B6F7R2 E9JD71 A0A1B6DW40 A0A232F3Y7 K7J0J3 A0A0C9RD96 A0A0L7RGV4 A0A154PQ91 T1I4G8 J9JPM5 A0A023F487 A0A1Y1L2K6 A0A2S2NQB3 A0A2S2R4K8 D6WDN2 A0A0C9PU86 A0A1W4X9P2 A0A224X8S2 A0A0A9WC65 A0A2P6L738 A0A182WHB2 A0A1B0CX19 A0A182M032 A0A182RG46 A0A182Q9P6 A0A084VY71 A0A182I5B6 Q7PVN1 A0A182VGL6 A0A182UAQ9 A0A182KU93 A0A182XKS9 A0A3L8DUU7 A0A182KC16 A0A2L2YET9 A0A0C9QW66 A0A182PAN0 A0A182Y9X1 U5ELH0 A0A182NF89 A0A182J4L7 T1J981 A0A1E1X548 A0A2M4AN54 A0A131YNT2 A0A224YY59 A0A1E1XMB8 W5JP59 B7Q7M8 A0A1B0DC22 L7M9U3 B0WB15 A0A182F121 L7LVG7 Q17LW5 A0A0P5KTU8 A0A0N8C9N4 A0A1W4UYR8 A0A0P5YWD9 A0A0P6IA13 A0A1I8NBP5 A0A0P5KII8 B4KI03 T1P995 A0A1A9XEP8 B3N3X1 E9H9Z7 B4P1Y7 A0A1A9WKK9 W8B5U8 A0A1B0FEC7 A0A1A9V886 A0A3B0K2J4 A0A034VW87 B4IE65 B4Q3Q3
Pubmed
19121390
26354079
26227816
22118469
24845553
21347285
+ More
24508170 20798317 21719571 20566863 21282665 28648823 20075255 25474469 28004739 18362917 19820115 25401762 24438588 12364791 14747013 17210077 20966253 30249741 26561354 25244985 28503490 26830274 28797301 29209593 20920257 23761445 25576852 17510324 25315136 17994087 21292972 17550304 24495485 25348373 22936249
24508170 20798317 21719571 20566863 21282665 28648823 20075255 25474469 28004739 18362917 19820115 25401762 24438588 12364791 14747013 17210077 20966253 30249741 26561354 25244985 28503490 26830274 28797301 29209593 20920257 23761445 25576852 17510324 25315136 17994087 21292972 17550304 24495485 25348373 22936249
EMBL
BABH01005321
NWSH01000232
PCG78163.1
ODYU01003798
SOQ43003.1
GDQN01008049
+ More
JAT83005.1 KQ461194 KPJ06912.1 KQ459603 KPI92920.1 JTDY01000095 KOB78901.1 AGBW02014975 OWR40794.1 NEVH01006721 PNF37142.1 KK852870 KDR14604.1 LBMM01004121 KMQ92796.1 ADTU01003771 ADTU01003772 KK107941 EZA47095.1 GL442175 EFN63835.1 KZ288269 PBC29972.1 KQ981768 KYN36003.1 KQ435809 KOX72969.1 KQ977811 KYM99513.1 GL888331 EGI62678.1 KQ980055 KYN17986.1 KQ760243 OAD61280.1 GL453975 EFN75102.1 DS235354 EEB15085.1 GECZ01023497 JAS46272.1 GL771866 EFZ09242.1 GEDC01007449 JAS29849.1 NNAY01001055 OXU25322.1 GBYB01004911 JAG74678.1 KQ414596 KOC70090.1 KQ435037 KZC14085.1 ACPB03008625 ABLF02018146 GBBI01002401 JAC16311.1 GEZM01068327 JAV67008.1 GGMR01006752 MBY19371.1 GGMS01015089 MBY84292.1 KQ971322 EEZ99946.1 GBYB01004913 JAG74680.1 GFTR01007701 JAW08725.1 GBHO01038626 GBHO01038625 GBHO01038624 GBHO01038623 GBRD01013515 JAG04978.1 JAG04979.1 JAG04980.1 JAG04981.1 JAG52311.1 MWRG01001338 PRD34371.1 AJWK01033131 AXCM01000590 AXCN02000824 ATLV01018323 KE525231 KFB42915.1 APCN01003844 AAAB01008984 EAA15037.3 QOIP01000004 RLU24056.1 IAAA01028742 IAAA01028743 IAAA01028744 LAA06666.1 GBYB01004912 JAG74679.1 GANO01001411 JAB58460.1 JH431970 GFAC01004788 JAT94400.1 GGFK01008905 MBW42226.1 GEDV01008339 JAP80218.1 GFPF01007606 MAA18752.1 GFAA01003215 JAU00220.1 ADMH02000504 ETN66172.1 ABJB010022040 DS876681 EEC14850.1 AJVK01013894 GACK01005145 JAA59889.1 DS231876 EDS41973.1 GACK01010136 JAA54898.1 CH477210 EAT47706.1 GDIQ01184756 GDIQ01053857 GDIQ01038806 LRGB01001361 JAK66969.1 KZS12595.1 GDIQ01101187 JAL50539.1 GDIP01053706 JAM50009.1 GDIQ01007480 JAN87257.1 GDIQ01190664 JAK61061.1 CH933807 EDW11285.1 KA644478 AFP59107.1 CH954177 EDV58823.1 GL732609 EFX71492.1 CM000157 EDW88158.1 GAMC01021361 JAB85194.1 CCAG010005473 OUUW01000004 SPP79826.1 GAKP01012842 JAC46110.1 CH480831 EDW45892.1 CM000361 CM002910 EDX04778.1 KMY89877.1
JAT83005.1 KQ461194 KPJ06912.1 KQ459603 KPI92920.1 JTDY01000095 KOB78901.1 AGBW02014975 OWR40794.1 NEVH01006721 PNF37142.1 KK852870 KDR14604.1 LBMM01004121 KMQ92796.1 ADTU01003771 ADTU01003772 KK107941 EZA47095.1 GL442175 EFN63835.1 KZ288269 PBC29972.1 KQ981768 KYN36003.1 KQ435809 KOX72969.1 KQ977811 KYM99513.1 GL888331 EGI62678.1 KQ980055 KYN17986.1 KQ760243 OAD61280.1 GL453975 EFN75102.1 DS235354 EEB15085.1 GECZ01023497 JAS46272.1 GL771866 EFZ09242.1 GEDC01007449 JAS29849.1 NNAY01001055 OXU25322.1 GBYB01004911 JAG74678.1 KQ414596 KOC70090.1 KQ435037 KZC14085.1 ACPB03008625 ABLF02018146 GBBI01002401 JAC16311.1 GEZM01068327 JAV67008.1 GGMR01006752 MBY19371.1 GGMS01015089 MBY84292.1 KQ971322 EEZ99946.1 GBYB01004913 JAG74680.1 GFTR01007701 JAW08725.1 GBHO01038626 GBHO01038625 GBHO01038624 GBHO01038623 GBRD01013515 JAG04978.1 JAG04979.1 JAG04980.1 JAG04981.1 JAG52311.1 MWRG01001338 PRD34371.1 AJWK01033131 AXCM01000590 AXCN02000824 ATLV01018323 KE525231 KFB42915.1 APCN01003844 AAAB01008984 EAA15037.3 QOIP01000004 RLU24056.1 IAAA01028742 IAAA01028743 IAAA01028744 LAA06666.1 GBYB01004912 JAG74679.1 GANO01001411 JAB58460.1 JH431970 GFAC01004788 JAT94400.1 GGFK01008905 MBW42226.1 GEDV01008339 JAP80218.1 GFPF01007606 MAA18752.1 GFAA01003215 JAU00220.1 ADMH02000504 ETN66172.1 ABJB010022040 DS876681 EEC14850.1 AJVK01013894 GACK01005145 JAA59889.1 DS231876 EDS41973.1 GACK01010136 JAA54898.1 CH477210 EAT47706.1 GDIQ01184756 GDIQ01053857 GDIQ01038806 LRGB01001361 JAK66969.1 KZS12595.1 GDIQ01101187 JAL50539.1 GDIP01053706 JAM50009.1 GDIQ01007480 JAN87257.1 GDIQ01190664 JAK61061.1 CH933807 EDW11285.1 KA644478 AFP59107.1 CH954177 EDV58823.1 GL732609 EFX71492.1 CM000157 EDW88158.1 GAMC01021361 JAB85194.1 CCAG010005473 OUUW01000004 SPP79826.1 GAKP01012842 JAC46110.1 CH480831 EDW45892.1 CM000361 CM002910 EDX04778.1 KMY89877.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000235965 UP000027135 UP000036403 UP000005205 UP000053097 UP000000311 UP000242457 UP000078541 UP000005203 UP000053105 UP000078542 UP000007755 UP000078492 UP000008237 UP000009046 UP000215335 UP000002358 UP000053825 UP000076502 UP000015103 UP000007819 UP000007266 UP000192223 UP000075920 UP000092461 UP000075883 UP000075900 UP000075886 UP000030765 UP000075840 UP000007062 UP000075903 UP000075902 UP000075882 UP000076407 UP000279307 UP000075881 UP000075885 UP000076408 UP000075884 UP000075880 UP000000673 UP000001555 UP000092462 UP000002320 UP000069272 UP000008820 UP000076858 UP000192221 UP000095301 UP000009192 UP000092443 UP000008711 UP000000305 UP000002282 UP000091820 UP000092444 UP000078200 UP000268350 UP000001292 UP000000304
UP000235965 UP000027135 UP000036403 UP000005205 UP000053097 UP000000311 UP000242457 UP000078541 UP000005203 UP000053105 UP000078542 UP000007755 UP000078492 UP000008237 UP000009046 UP000215335 UP000002358 UP000053825 UP000076502 UP000015103 UP000007819 UP000007266 UP000192223 UP000075920 UP000092461 UP000075883 UP000075900 UP000075886 UP000030765 UP000075840 UP000007062 UP000075903 UP000075902 UP000075882 UP000076407 UP000279307 UP000075881 UP000075885 UP000076408 UP000075884 UP000075880 UP000000673 UP000001555 UP000092462 UP000002320 UP000069272 UP000008820 UP000076858 UP000192221 UP000095301 UP000009192 UP000092443 UP000008711 UP000000305 UP000002282 UP000091820 UP000092444 UP000078200 UP000268350 UP000001292 UP000000304
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JLX2
A0A2A4K309
A0A2H1VQ83
A0A1E1W7S9
A0A194QN25
A0A194PP61
+ More
A0A0L7LUK9 A0A212EH40 A0A2J7R8I2 A0A067QYM9 A0A0J7KR76 A0A158NYE5 A0A026VTW0 E2ARV2 A0A2A3EFB4 A0A195F6A6 A0A087ZYU3 A0A0M8ZZY0 A0A151IEZ9 F4WT65 A0A151J547 A0A310SUR3 E2CAD6 E0VNX9 A0A1B6F7R2 E9JD71 A0A1B6DW40 A0A232F3Y7 K7J0J3 A0A0C9RD96 A0A0L7RGV4 A0A154PQ91 T1I4G8 J9JPM5 A0A023F487 A0A1Y1L2K6 A0A2S2NQB3 A0A2S2R4K8 D6WDN2 A0A0C9PU86 A0A1W4X9P2 A0A224X8S2 A0A0A9WC65 A0A2P6L738 A0A182WHB2 A0A1B0CX19 A0A182M032 A0A182RG46 A0A182Q9P6 A0A084VY71 A0A182I5B6 Q7PVN1 A0A182VGL6 A0A182UAQ9 A0A182KU93 A0A182XKS9 A0A3L8DUU7 A0A182KC16 A0A2L2YET9 A0A0C9QW66 A0A182PAN0 A0A182Y9X1 U5ELH0 A0A182NF89 A0A182J4L7 T1J981 A0A1E1X548 A0A2M4AN54 A0A131YNT2 A0A224YY59 A0A1E1XMB8 W5JP59 B7Q7M8 A0A1B0DC22 L7M9U3 B0WB15 A0A182F121 L7LVG7 Q17LW5 A0A0P5KTU8 A0A0N8C9N4 A0A1W4UYR8 A0A0P5YWD9 A0A0P6IA13 A0A1I8NBP5 A0A0P5KII8 B4KI03 T1P995 A0A1A9XEP8 B3N3X1 E9H9Z7 B4P1Y7 A0A1A9WKK9 W8B5U8 A0A1B0FEC7 A0A1A9V886 A0A3B0K2J4 A0A034VW87 B4IE65 B4Q3Q3
A0A0L7LUK9 A0A212EH40 A0A2J7R8I2 A0A067QYM9 A0A0J7KR76 A0A158NYE5 A0A026VTW0 E2ARV2 A0A2A3EFB4 A0A195F6A6 A0A087ZYU3 A0A0M8ZZY0 A0A151IEZ9 F4WT65 A0A151J547 A0A310SUR3 E2CAD6 E0VNX9 A0A1B6F7R2 E9JD71 A0A1B6DW40 A0A232F3Y7 K7J0J3 A0A0C9RD96 A0A0L7RGV4 A0A154PQ91 T1I4G8 J9JPM5 A0A023F487 A0A1Y1L2K6 A0A2S2NQB3 A0A2S2R4K8 D6WDN2 A0A0C9PU86 A0A1W4X9P2 A0A224X8S2 A0A0A9WC65 A0A2P6L738 A0A182WHB2 A0A1B0CX19 A0A182M032 A0A182RG46 A0A182Q9P6 A0A084VY71 A0A182I5B6 Q7PVN1 A0A182VGL6 A0A182UAQ9 A0A182KU93 A0A182XKS9 A0A3L8DUU7 A0A182KC16 A0A2L2YET9 A0A0C9QW66 A0A182PAN0 A0A182Y9X1 U5ELH0 A0A182NF89 A0A182J4L7 T1J981 A0A1E1X548 A0A2M4AN54 A0A131YNT2 A0A224YY59 A0A1E1XMB8 W5JP59 B7Q7M8 A0A1B0DC22 L7M9U3 B0WB15 A0A182F121 L7LVG7 Q17LW5 A0A0P5KTU8 A0A0N8C9N4 A0A1W4UYR8 A0A0P5YWD9 A0A0P6IA13 A0A1I8NBP5 A0A0P5KII8 B4KI03 T1P995 A0A1A9XEP8 B3N3X1 E9H9Z7 B4P1Y7 A0A1A9WKK9 W8B5U8 A0A1B0FEC7 A0A1A9V886 A0A3B0K2J4 A0A034VW87 B4IE65 B4Q3Q3
PDB
6IE1
E-value=4.80795e-113,
Score=1046
Ontologies
GO
Topology
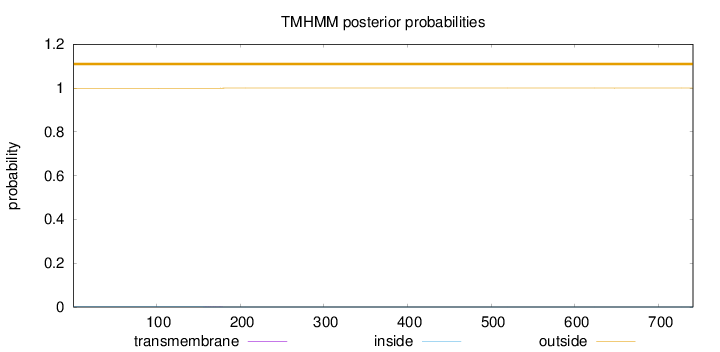
Length:
741
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01403
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00073
outside
1 - 741
Population Genetic Test Statistics
Pi
194.537188
Theta
234.157085
Tajima's D
-0.9194
CLR
15.470509
CSRT
0.158542072896355
Interpretation
Uncertain
