Gene
KWMTBOMO07074
Pre Gene Modal
BGIBMGA010436
Annotation
PREDICTED:_spondin-1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.663
Sequence
CDS
ATGAAGTCGATTTGGAGAGCGTTCTTATTGTTACACTATTTAAATTTTCATAAAATTATGTGTTTCCGTTGCGATCGCCGCCCTTATGGCTCTACAGCGCCAGCTTCACCACCGGATGGTCGTTTTCATCTCAATGTACTTGGAGTTGAAAATCATGCTTACTTACCAGAACAAGTCTATACAGCACAAATAACGTCGACCGATGGCGAATCCAAATTTACGGGTTTCATGATATCAGCTGAGGGGGACACTCGACAAGACCCGAAGAACCCACGGAGACTCATCTCCTTATTCCCGGGTGATATTAGGCCGCAGAAAATCGAAACGGCTAAGTTTAGTGACCGCTGTCTCTACTCAGTCGAGCAAGCTACGCGCACTTGGAAAACTTCTGTTGAGGTGTTTTGGCAAGCCCCACCCACAGGTCACGGTTGTGTTACATTGCGTGCTGTGGTAGTTGAAAACGATGAACAGTGGTTCGAAGATGGTGCTCCGCTCACGCAAAAGATCTGCGAGGACTTAAGGCAGCCCGACGACATGTCGCCACTTATAAATTACGATTGTCAGATCTGTGATGAAGCTAAATATGAGATATCTTTCACCGGCGTTTGGTCCCGTAATACTCATCCTCGATTATATCCTGAAAACGTTTGGGTCCCTAGATTTAGTGATTTAGTTGGCGCTTCTCACGATGTGGACTATTTAATGTGGATACCGGGCAACTTGGCCGGTGAAGGAATGCGGGACTTAGCCGAACATGCTAATTCGAGTAAACTCGAAACCGAAATAAGAGAAAAGATAGGGGACGGGGTACGTACTCTCATTAAGGGTAAAGGCCACGGCTATCGCAAAATGAACAATCCAACGTTCGCGATTTTTCGCGCGGATAAAAAAAACCATTTAGTGTCGGTTGCGGTGGCTCTTCATCCATCACCAGATTGGTTTCTTGGAGTTACGAGGTTTGAACTGTGTCAGGAAGATAATACTTGGCTACGGGAGAGAGAGTTGAATCTCTTTCCATGGGATGCTGGGACAGACAGCGGTGTATCGTATGAGTCACCGAACATTGAAACGTATCCGCAAGACACGATATCTCGCGTTGCAATGAGTTCATACGACAAAAAATCGCCCTTCTACCAAACGACAATGACGGAAATGCATCCGTTCGGAAAACTCCACATCAAATTCATAAGGAGCTATCACAGAGAATGCGAAGACGAGGAAGAAACTACAGAAACGACGAGTCCTGAAGGAGATCTAGAAAGTACATCAACCACAGAGACACCTACTACGGAAGAGTCGGAAGGACCTTTCGAGCCAAAGCCGCCATCCAGCGAAGATCCGATAGTATTCGATCAGGATTCTTCTGAAGAATGTCCCATGTCTGAATGGCAAGCCTGGAGTTCATGTGAAGGGCCCTGCGACCACGGACACGTCAAGGGGTACCAATGGCGGGTGAGGTACCATCTCGTCGATGGCATTGCCGTTGAGAAATACGATCCTTATAATCAGCACACTTTAAATCTAGAAGTGCCAAAGTATTGCAAAAATCACGTCGAAGATTTTCAAAGGAAAGAGTGCTTCGAGAAATGCTCTGAAGATTACGATGAGCATGAAGACGAAATAAAAATGGAGAGAAGGATAATATCGGAGATCATGCCTGGTTCTTCATGGGGTGGATCGAGTAGAAAAAATACCAAGCATTAA
Protein
MKSIWRAFLLLHYLNFHKIMCFRCDRRPYGSTAPASPPDGRFHLNVLGVENHAYLPEQVYTAQITSTDGESKFTGFMISAEGDTRQDPKNPRRLISLFPGDIRPQKIETAKFSDRCLYSVEQATRTWKTSVEVFWQAPPTGHGCVTLRAVVVENDEQWFEDGAPLTQKICEDLRQPDDMSPLINYDCQICDEAKYEISFTGVWSRNTHPRLYPENVWVPRFSDLVGASHDVDYLMWIPGNLAGEGMRDLAEHANSSKLETEIREKIGDGVRTLIKGKGHGYRKMNNPTFAIFRADKKNHLVSVAVALHPSPDWFLGVTRFELCQEDNTWLRERELNLFPWDAGTDSGVSYESPNIETYPQDTISRVAMSSYDKKSPFYQTTMTEMHPFGKLHIKFIRSYHRECEDEEETTETTSPEGDLESTSTTETPTTEESEGPFEPKPPSSEDPIVFDQDSSEECPMSEWQAWSSCEGPCDHGHVKGYQWRVRYHLVDGIAVEKYDPYNQHTLNLEVPKYCKNHVEDFQRKECFEKCSEDYDEHEDEIKMERRIISEIMPGSSWGGSSRKNTKH
Summary
Uniprot
H9JLN5
A0A2H1VRZ9
A0A3S2PHU8
A0A212EH32
A0A2A4K2N3
A0A2A4K1M3
+ More
A0A3S2NIG3 A0A194PHX8 A0A2H1VQ85 U5EYT8 Q170L4 A0A023F2I8 A0A182M9X5 A0A182W944 A0A182R5C7 A0A182Y1R5 A0A182PBS9 A0A2M4CJ21 A0A182JBS3 A0A0P5PL34 A0A1Q3FPJ2 A0A1Q3FP78 A0A2J7PR62 A0A1B6JTC9 A0A1J1IVV5 A0A0P5R1Y0 A0A224XEU1 A0A2J7PR48 A0A2J7PR47 A0A0P6FLY3 A0A2J7PR63 A0A0P5EHT2 A0A0P5A418 A0A0P6FNM0 A0A0P5H4F8 A0A0P5SZF3 A0A0P5EMH8 A0A0N8CXC5 A0A0P5Z9I4 A0A0P5EU30 A0A0P5AL01 A0A0P4ZG86 A0A0P5QQF6 A0A0N8CEJ0 A0A0P5PPB9 A0A0P5WQD1 A0A0N8ARA9 A0A0P5MPN1 A0A0P6GS18 A0A0P6DCY0 A0A0P5I8X5 A0A0P5CIT8 A0A0P5SCA1 A0A0N8AJJ8 A0A0P5TWQ4 A0A0P5G5S6 A0A0N8BMI8 A0A0P5K323 A0A0N8BPV7 A0A0P5S9E5 A0A0P6EAL7 A0A0P5UAZ6 A0A0P5E4B7 A0A0P5MWG3 A0A0P5XD67 A0A0P5UGB3 A0A0P5T3I4 A0A336LL51 A0A0P6CTJ0 A0A0P5IU67 A0A0P5KVB3 A0A0P6I4P9 A0A0P5KSU2 A0A0P5TFB0 E0VCJ0 A0A164MD69 E2BUL2 A0A2P8YCY1
A0A3S2NIG3 A0A194PHX8 A0A2H1VQ85 U5EYT8 Q170L4 A0A023F2I8 A0A182M9X5 A0A182W944 A0A182R5C7 A0A182Y1R5 A0A182PBS9 A0A2M4CJ21 A0A182JBS3 A0A0P5PL34 A0A1Q3FPJ2 A0A1Q3FP78 A0A2J7PR62 A0A1B6JTC9 A0A1J1IVV5 A0A0P5R1Y0 A0A224XEU1 A0A2J7PR48 A0A2J7PR47 A0A0P6FLY3 A0A2J7PR63 A0A0P5EHT2 A0A0P5A418 A0A0P6FNM0 A0A0P5H4F8 A0A0P5SZF3 A0A0P5EMH8 A0A0N8CXC5 A0A0P5Z9I4 A0A0P5EU30 A0A0P5AL01 A0A0P4ZG86 A0A0P5QQF6 A0A0N8CEJ0 A0A0P5PPB9 A0A0P5WQD1 A0A0N8ARA9 A0A0P5MPN1 A0A0P6GS18 A0A0P6DCY0 A0A0P5I8X5 A0A0P5CIT8 A0A0P5SCA1 A0A0N8AJJ8 A0A0P5TWQ4 A0A0P5G5S6 A0A0N8BMI8 A0A0P5K323 A0A0N8BPV7 A0A0P5S9E5 A0A0P6EAL7 A0A0P5UAZ6 A0A0P5E4B7 A0A0P5MWG3 A0A0P5XD67 A0A0P5UGB3 A0A0P5T3I4 A0A336LL51 A0A0P6CTJ0 A0A0P5IU67 A0A0P5KVB3 A0A0P6I4P9 A0A0P5KSU2 A0A0P5TFB0 E0VCJ0 A0A164MD69 E2BUL2 A0A2P8YCY1
EMBL
BABH01005319
BABH01005320
BABH01005321
ODYU01003798
SOQ43004.1
RSAL01000028
+ More
RVE51860.1 AGBW02014975 OWR40793.1 NWSH01000232 PCG78164.1 PCG78165.1 RVE51861.1 KQ459603 KPI92922.1 SOQ43005.1 GANO01000077 JAB59794.1 CH477472 EAT40381.1 GBBI01003473 JAC15239.1 AXCM01008352 GGFL01000700 MBW64878.1 GDIQ01132338 JAL19388.1 GFDL01005525 JAV29520.1 GFDL01005644 JAV29401.1 NEVH01022633 PNF18806.1 GECU01005533 JAT02174.1 CVRI01000059 CRL03842.1 GDIQ01108980 JAL42746.1 GFTR01008128 JAW08298.1 PNF18808.1 PNF18807.1 GDIQ01045511 JAN49226.1 PNF18809.1 GDIQ01270118 JAJ81606.1 GDIP01208772 JAJ14630.1 GDIQ01045510 JAN49227.1 GDIQ01233233 GDIQ01119073 GDIP01095690 JAK18492.1 JAM08025.1 GDIP01134289 JAL69425.1 GDIQ01267301 JAJ84423.1 GDIP01089647 JAM14068.1 GDIP01046852 JAM56863.1 GDIQ01267300 GDIQ01197652 JAJ84424.1 GDIP01197830 JAJ25572.1 GDIP01213072 JAJ10330.1 GDIQ01113789 JAL37937.1 GDIP01139527 JAL64187.1 GDIQ01125376 JAL26350.1 GDIP01095689 JAM08026.1 GDIQ01250587 GDIQ01233232 GDIQ01163152 GDIQ01132337 JAK01138.1 GDIQ01156604 JAK95121.1 GDIQ01029118 JAN65619.1 GDIQ01079590 GDIQ01048851 JAN15147.1 GDIQ01217146 JAK34579.1 GDIP01185568 JAJ37834.1 GDIP01141635 JAL62079.1 GDIQ01269475 JAJ82249.1 GDIP01138432 JAL65282.1 GDIQ01267746 JAJ83978.1 GDIQ01265527 GDIQ01233231 GDIQ01203000 GDIQ01188747 GDIQ01163155 GDIQ01160359 GDIQ01119072 JAK88570.1 GDIQ01215998 GDIQ01112476 JAK35727.1 GDIQ01156603 JAK95122.1 GDIP01142732 JAL60982.1 GDIQ01065551 JAN29186.1 GDIP01132845 JAL70869.1 GDIP01147227 JAJ76175.1 GDIQ01158613 JAK93112.1 GDIP01073851 JAM29864.1 GDIP01113440 JAL90274.1 GDIP01131532 JAL72182.1 UFQT01000038 SSX18435.1 GDIQ01087539 JAN07198.1 GDIQ01210190 JAK41535.1 GDIQ01180097 JAK71628.1 GDIQ01009616 JAN85121.1 GDIQ01180096 JAK71629.1 GDIP01127234 JAL76480.1 DS235053 EEB11096.1 LRGB01003024 KZS04951.1 GL450721 EFN80621.1 PYGN01000696 PSN42107.1
RVE51860.1 AGBW02014975 OWR40793.1 NWSH01000232 PCG78164.1 PCG78165.1 RVE51861.1 KQ459603 KPI92922.1 SOQ43005.1 GANO01000077 JAB59794.1 CH477472 EAT40381.1 GBBI01003473 JAC15239.1 AXCM01008352 GGFL01000700 MBW64878.1 GDIQ01132338 JAL19388.1 GFDL01005525 JAV29520.1 GFDL01005644 JAV29401.1 NEVH01022633 PNF18806.1 GECU01005533 JAT02174.1 CVRI01000059 CRL03842.1 GDIQ01108980 JAL42746.1 GFTR01008128 JAW08298.1 PNF18808.1 PNF18807.1 GDIQ01045511 JAN49226.1 PNF18809.1 GDIQ01270118 JAJ81606.1 GDIP01208772 JAJ14630.1 GDIQ01045510 JAN49227.1 GDIQ01233233 GDIQ01119073 GDIP01095690 JAK18492.1 JAM08025.1 GDIP01134289 JAL69425.1 GDIQ01267301 JAJ84423.1 GDIP01089647 JAM14068.1 GDIP01046852 JAM56863.1 GDIQ01267300 GDIQ01197652 JAJ84424.1 GDIP01197830 JAJ25572.1 GDIP01213072 JAJ10330.1 GDIQ01113789 JAL37937.1 GDIP01139527 JAL64187.1 GDIQ01125376 JAL26350.1 GDIP01095689 JAM08026.1 GDIQ01250587 GDIQ01233232 GDIQ01163152 GDIQ01132337 JAK01138.1 GDIQ01156604 JAK95121.1 GDIQ01029118 JAN65619.1 GDIQ01079590 GDIQ01048851 JAN15147.1 GDIQ01217146 JAK34579.1 GDIP01185568 JAJ37834.1 GDIP01141635 JAL62079.1 GDIQ01269475 JAJ82249.1 GDIP01138432 JAL65282.1 GDIQ01267746 JAJ83978.1 GDIQ01265527 GDIQ01233231 GDIQ01203000 GDIQ01188747 GDIQ01163155 GDIQ01160359 GDIQ01119072 JAK88570.1 GDIQ01215998 GDIQ01112476 JAK35727.1 GDIQ01156603 JAK95122.1 GDIP01142732 JAL60982.1 GDIQ01065551 JAN29186.1 GDIP01132845 JAL70869.1 GDIP01147227 JAJ76175.1 GDIQ01158613 JAK93112.1 GDIP01073851 JAM29864.1 GDIP01113440 JAL90274.1 GDIP01131532 JAL72182.1 UFQT01000038 SSX18435.1 GDIQ01087539 JAN07198.1 GDIQ01210190 JAK41535.1 GDIQ01180097 JAK71628.1 GDIQ01009616 JAN85121.1 GDIQ01180096 JAK71629.1 GDIP01127234 JAL76480.1 DS235053 EEB11096.1 LRGB01003024 KZS04951.1 GL450721 EFN80621.1 PYGN01000696 PSN42107.1
Proteomes
PRIDE
Interpro
ProteinModelPortal
H9JLN5
A0A2H1VRZ9
A0A3S2PHU8
A0A212EH32
A0A2A4K2N3
A0A2A4K1M3
+ More
A0A3S2NIG3 A0A194PHX8 A0A2H1VQ85 U5EYT8 Q170L4 A0A023F2I8 A0A182M9X5 A0A182W944 A0A182R5C7 A0A182Y1R5 A0A182PBS9 A0A2M4CJ21 A0A182JBS3 A0A0P5PL34 A0A1Q3FPJ2 A0A1Q3FP78 A0A2J7PR62 A0A1B6JTC9 A0A1J1IVV5 A0A0P5R1Y0 A0A224XEU1 A0A2J7PR48 A0A2J7PR47 A0A0P6FLY3 A0A2J7PR63 A0A0P5EHT2 A0A0P5A418 A0A0P6FNM0 A0A0P5H4F8 A0A0P5SZF3 A0A0P5EMH8 A0A0N8CXC5 A0A0P5Z9I4 A0A0P5EU30 A0A0P5AL01 A0A0P4ZG86 A0A0P5QQF6 A0A0N8CEJ0 A0A0P5PPB9 A0A0P5WQD1 A0A0N8ARA9 A0A0P5MPN1 A0A0P6GS18 A0A0P6DCY0 A0A0P5I8X5 A0A0P5CIT8 A0A0P5SCA1 A0A0N8AJJ8 A0A0P5TWQ4 A0A0P5G5S6 A0A0N8BMI8 A0A0P5K323 A0A0N8BPV7 A0A0P5S9E5 A0A0P6EAL7 A0A0P5UAZ6 A0A0P5E4B7 A0A0P5MWG3 A0A0P5XD67 A0A0P5UGB3 A0A0P5T3I4 A0A336LL51 A0A0P6CTJ0 A0A0P5IU67 A0A0P5KVB3 A0A0P6I4P9 A0A0P5KSU2 A0A0P5TFB0 E0VCJ0 A0A164MD69 E2BUL2 A0A2P8YCY1
A0A3S2NIG3 A0A194PHX8 A0A2H1VQ85 U5EYT8 Q170L4 A0A023F2I8 A0A182M9X5 A0A182W944 A0A182R5C7 A0A182Y1R5 A0A182PBS9 A0A2M4CJ21 A0A182JBS3 A0A0P5PL34 A0A1Q3FPJ2 A0A1Q3FP78 A0A2J7PR62 A0A1B6JTC9 A0A1J1IVV5 A0A0P5R1Y0 A0A224XEU1 A0A2J7PR48 A0A2J7PR47 A0A0P6FLY3 A0A2J7PR63 A0A0P5EHT2 A0A0P5A418 A0A0P6FNM0 A0A0P5H4F8 A0A0P5SZF3 A0A0P5EMH8 A0A0N8CXC5 A0A0P5Z9I4 A0A0P5EU30 A0A0P5AL01 A0A0P4ZG86 A0A0P5QQF6 A0A0N8CEJ0 A0A0P5PPB9 A0A0P5WQD1 A0A0N8ARA9 A0A0P5MPN1 A0A0P6GS18 A0A0P6DCY0 A0A0P5I8X5 A0A0P5CIT8 A0A0P5SCA1 A0A0N8AJJ8 A0A0P5TWQ4 A0A0P5G5S6 A0A0N8BMI8 A0A0P5K323 A0A0N8BPV7 A0A0P5S9E5 A0A0P6EAL7 A0A0P5UAZ6 A0A0P5E4B7 A0A0P5MWG3 A0A0P5XD67 A0A0P5UGB3 A0A0P5T3I4 A0A336LL51 A0A0P6CTJ0 A0A0P5IU67 A0A0P5KVB3 A0A0P6I4P9 A0A0P5KSU2 A0A0P5TFB0 E0VCJ0 A0A164MD69 E2BUL2 A0A2P8YCY1
PDB
3Q13
E-value=3.38562e-33,
Score=356
Ontologies
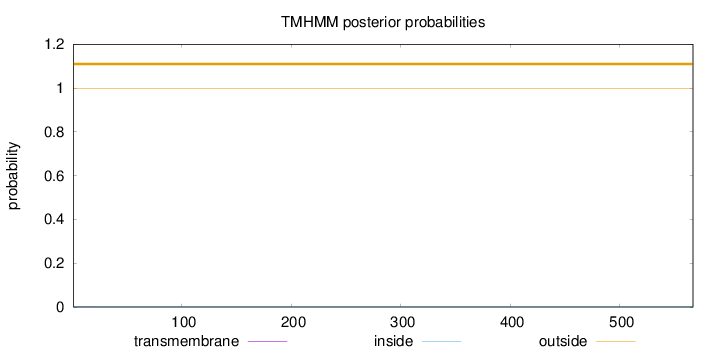
Topology
Length:
567
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0046
Exp number, first 60 AAs:
0.00419
Total prob of N-in:
0.00094
outside
1 - 567
Population Genetic Test Statistics
Pi
205.70304
Theta
169.506872
Tajima's D
0.663824
CLR
0.126192
CSRT
0.56067196640168
Interpretation
Uncertain
