Gene
KWMTBOMO07073
Pre Gene Modal
BGIBMGA010436
Annotation
PREDICTED:_spondin-2-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.464
Sequence
CDS
ATGCATTCGAAGAAGTTATTCGGTATTCGTGGAGTTGTTTTCCTGACGTTATTGTTCGGTTTAAGTGTTTGCGATGAAGACGTCTGCAATCGGCGACCGCTCGGGACTAAGACGGATCCGCTACCACCTGACAATCAGTTCCTCATTGAAATAAACGGATTAATTGAAAACCTTTATATACCGAAGAAAGAGTACATAGTACGTTTGAGATCGAGAGATGCCAAATCAACTTTCATAGCGTTTACGATAGCACTGAAAGAGGACACGCTGCCGAATAGGAACAACCCCCGGAAACCGATCCATTTGAACGCTGGACGTTTGACGCTGATGCCGAACTCTACCACGACTAGATACAAGCAGCATTGCGATGATACTGTTGTCGAATTGGATATAACGCCGAAAACTTTTGTGGAGGCACTTTGGACGGCTCCGGACAAGGACAACAAATGCGTTACAGTTTTCGCGGTGGTAGCTGTACAACCCGATGTCTGGTATAATTACGAAGGTCCGCTGTCAAAATGGATCTGCGAAGATAGGCGAAAGGCTGACGATATACAGCCTTTAGAAAATGACAACTGCCAAGCTTGTGAAGATGCAAGGTACCAGTTGACCTTCGAAGGTACATGGTCGTACAACACGCATCCATCGCTGTTTCCGAAATCCCGGGAACTGGCCCGGTTCAGTGACGTAGTCGGAGCATCGCACAGCAAAGACTTCAGCCTTTACAAATATACTTCTGAAGCTAGTTTAGGCTTGAAATTGTTGGCCGAACAAGGGAATAGTACTAAATTAGAGATCGAAATACAAAAACAGCTAGGTACATCTGTACGGACCGTGATCAAGGCATCAGCTCCTTACAACACAAGCATGTCAACGGTCACGAATTTTCGGGCTACGAGAGAACACCATCTAATTTCATTGGCAACGGCGTTGCTTCCATCGCCGGACTGGTTCCTCGGAGTCGCGAATTTAGAGTTATGCAGCATCAAGACCAATCAATGGACCGAAAACATAACTTTCAACTTGTATCCAATGGATGCTGGAACTGATAAGGGTTTGACTTTTGAGGCACCGAACGAGGAAATGTCCCCGCCACAGCCTATTTCTCCTGCGAAGTTCATTAAAGAAATACCAAAAGAGTTGATGAAGCCATTCGCTAGACTTAGATTGAAGCTGGTTCGAACATATAGTACTACTAATTGTTCCATGGAAATTGAGGAACCAGTCACAAGCGAAGGTGATAATGATGCTGATGCCGGAAATGACAAAGATAGAGAGCATAATTATATAACTGATCCTCCAGTGACCACTCCGCCGACAACAACGACAAGCGAAGAGCCACCGTCCGTCGATCCGGCATCACCGGATGACTGTCCACTATCAACGTGGGAAGAATGGTTTCCTTGCCAAGGAGTATGTAACGACGGGAAAATCAAAGGAATACAAATACGATTGCGATACCGCATGGTAGATGGTGTAGCTGTTGGAAAATATTTCGGTGAGGAACAAAAAGAATCACAAACACCAGAGGAATGTTTAGATAAATACAGTCTTAGCGACACTAGGGAATGTGAGGAAACATGCGATAACGAATCGGAAGTTGACGAAGATAACGTTTGGGATGACACAACATCCGAACTTGACGAACAAGCTGTAAAATAA
Protein
MHSKKLFGIRGVVFLTLLFGLSVCDEDVCNRRPLGTKTDPLPPDNQFLIEINGLIENLYIPKKEYIVRLRSRDAKSTFIAFTIALKEDTLPNRNNPRKPIHLNAGRLTLMPNSTTTRYKQHCDDTVVELDITPKTFVEALWTAPDKDNKCVTVFAVVAVQPDVWYNYEGPLSKWICEDRRKADDIQPLENDNCQACEDARYQLTFEGTWSYNTHPSLFPKSRELARFSDVVGASHSKDFSLYKYTSEASLGLKLLAEQGNSTKLEIEIQKQLGTSVRTVIKASAPYNTSMSTVTNFRATREHHLISLATALLPSPDWFLGVANLELCSIKTNQWTENITFNLYPMDAGTDKGLTFEAPNEEMSPPQPISPAKFIKEIPKELMKPFARLRLKLVRTYSTTNCSMEIEEPVTSEGDNDADAGNDKDREHNYITDPPVTTPPTTTTSEEPPSVDPASPDDCPLSTWEEWFPCQGVCNDGKIKGIQIRLRYRMVDGVAVGKYFGEEQKESQTPEECLDKYSLSDTRECEETCDNESEVDEDNVWDDTTSELDEQAVK
Summary
Uniprot
H9JLN5
A0A2H1VQ85
A0A2A4K1M3
A0A3S2NIG3
A0A194PHX8
A0A194QTN9
+ More
A0A0L7LTX0 A0A2H1VRZ9 A0A3S2PHU8 A0A212EH32 A0A182MYL4 A0A1B6F720 A0A1L8E3B7 A0A1L8E3B6 A0A1B6JTC9 A0A182Y1R5 A0A182R5C7 A0A182JZY3 A0A182PBS9 A0A2M4CJ21 A0A182W944 A0A182M9X5 A0A182FU59 A0A182QL14 A0A182HQ46 A0A182UWY8 A0A182TS53 A0A182KSL1 A0A182JBS3 A0A182XNC5 A0A1B0EWS7 A0A1B6G9H8 Q170L4 A0A1Q3FPJ2 A0A1Q3FP78 U5EYT8 A0A0P4W445 A0A023F2I8 A0A2J7PR62 Q7Q082 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A0P4VWU6 A0A0P4W243 A0A224XEU1 V5GQI0 A0A336LL51 A0A1J1IVV5 A0A2P2HWJ3 A0A131YJI2 A0A224Y7S0 A0A147BHW8 A0A1Y1JWI6 A0A1W4U6S0
A0A0L7LTX0 A0A2H1VRZ9 A0A3S2PHU8 A0A212EH32 A0A182MYL4 A0A1B6F720 A0A1L8E3B7 A0A1L8E3B6 A0A1B6JTC9 A0A182Y1R5 A0A182R5C7 A0A182JZY3 A0A182PBS9 A0A2M4CJ21 A0A182W944 A0A182M9X5 A0A182FU59 A0A182QL14 A0A182HQ46 A0A182UWY8 A0A182TS53 A0A182KSL1 A0A182JBS3 A0A182XNC5 A0A1B0EWS7 A0A1B6G9H8 Q170L4 A0A1Q3FPJ2 A0A1Q3FP78 U5EYT8 A0A0P4W445 A0A023F2I8 A0A2J7PR62 Q7Q082 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A0P4VWU6 A0A0P4W243 A0A224XEU1 V5GQI0 A0A336LL51 A0A1J1IVV5 A0A2P2HWJ3 A0A131YJI2 A0A224Y7S0 A0A147BHW8 A0A1Y1JWI6 A0A1W4U6S0
Pubmed
EMBL
BABH01005319
BABH01005320
BABH01005321
ODYU01003798
SOQ43005.1
NWSH01000232
+ More
PCG78165.1 RSAL01000028 RVE51861.1 KQ459603 KPI92922.1 KQ461194 KPJ06911.1 JTDY01000095 KOB78897.1 SOQ43004.1 RVE51860.1 AGBW02014975 OWR40793.1 GECZ01023720 JAS46049.1 GFDF01000867 JAV13217.1 GFDF01000866 JAV13218.1 GECU01005533 JAT02174.1 GGFL01000700 MBW64878.1 AXCM01008352 AXCN02001040 APCN01003292 AJWK01006547 AJWK01006548 AJWK01006549 GECZ01010684 JAS59085.1 CH477472 EAT40381.1 GFDL01005525 JAV29520.1 GFDL01005644 JAV29401.1 GANO01000077 JAB59794.1 GDRN01090813 JAI60406.1 GBBI01003473 JAC15239.1 NEVH01022633 PNF18806.1 AAAB01008986 EAA00346.3 PNF18807.1 PNF18808.1 PNF18809.1 GDRN01090815 JAI60405.1 GDRN01090812 JAI60407.1 GFTR01008128 JAW08298.1 GALX01002067 JAB66399.1 UFQT01000038 SSX18435.1 CVRI01000059 CRL03842.1 IACF01000366 LAB66149.1 GEDV01009957 JAP78600.1 GFPF01000413 MAA11559.1 GEGO01005005 JAR90399.1 GEZM01102876 JAV51755.1
PCG78165.1 RSAL01000028 RVE51861.1 KQ459603 KPI92922.1 KQ461194 KPJ06911.1 JTDY01000095 KOB78897.1 SOQ43004.1 RVE51860.1 AGBW02014975 OWR40793.1 GECZ01023720 JAS46049.1 GFDF01000867 JAV13217.1 GFDF01000866 JAV13218.1 GECU01005533 JAT02174.1 GGFL01000700 MBW64878.1 AXCM01008352 AXCN02001040 APCN01003292 AJWK01006547 AJWK01006548 AJWK01006549 GECZ01010684 JAS59085.1 CH477472 EAT40381.1 GFDL01005525 JAV29520.1 GFDL01005644 JAV29401.1 GANO01000077 JAB59794.1 GDRN01090813 JAI60406.1 GBBI01003473 JAC15239.1 NEVH01022633 PNF18806.1 AAAB01008986 EAA00346.3 PNF18807.1 PNF18808.1 PNF18809.1 GDRN01090815 JAI60405.1 GDRN01090812 JAI60407.1 GFTR01008128 JAW08298.1 GALX01002067 JAB66399.1 UFQT01000038 SSX18435.1 CVRI01000059 CRL03842.1 IACF01000366 LAB66149.1 GEDV01009957 JAP78600.1 GFPF01000413 MAA11559.1 GEGO01005005 JAR90399.1 GEZM01102876 JAV51755.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000037510
+ More
UP000007151 UP000075884 UP000076408 UP000075900 UP000075881 UP000075885 UP000075920 UP000075883 UP000069272 UP000075886 UP000075840 UP000075903 UP000075902 UP000075882 UP000075880 UP000076407 UP000092461 UP000008820 UP000235965 UP000007062 UP000183832 UP000192221
UP000007151 UP000075884 UP000076408 UP000075900 UP000075881 UP000075885 UP000075920 UP000075883 UP000069272 UP000075886 UP000075840 UP000075903 UP000075902 UP000075882 UP000075880 UP000076407 UP000092461 UP000008820 UP000235965 UP000007062 UP000183832 UP000192221
PRIDE
Pfam
Interpro
IPR042307
Reeler_sf
+ More
IPR038678 Spondin_N_sf
IPR002861 Reeler_dom
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR009465 Spondin_N
IPR038397 TBCC_N_sf
IPR017901 C-CAP_CF_C-like
IPR012945 Tubulin-bd_cofactor_C_dom
IPR031925 TBCC_N
IPR006599 CARP_motif
IPR016098 CAP/MinC_C
IPR002223 Kunitz_BPTI
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
IPR038678 Spondin_N_sf
IPR002861 Reeler_dom
IPR000884 TSP1_rpt
IPR036383 TSP1_rpt_sf
IPR009465 Spondin_N
IPR038397 TBCC_N_sf
IPR017901 C-CAP_CF_C-like
IPR012945 Tubulin-bd_cofactor_C_dom
IPR031925 TBCC_N
IPR006599 CARP_motif
IPR016098 CAP/MinC_C
IPR002223 Kunitz_BPTI
IPR036880 Kunitz_BPTI_sf
IPR020901 Prtase_inh_Kunz-CS
ProteinModelPortal
H9JLN5
A0A2H1VQ85
A0A2A4K1M3
A0A3S2NIG3
A0A194PHX8
A0A194QTN9
+ More
A0A0L7LTX0 A0A2H1VRZ9 A0A3S2PHU8 A0A212EH32 A0A182MYL4 A0A1B6F720 A0A1L8E3B7 A0A1L8E3B6 A0A1B6JTC9 A0A182Y1R5 A0A182R5C7 A0A182JZY3 A0A182PBS9 A0A2M4CJ21 A0A182W944 A0A182M9X5 A0A182FU59 A0A182QL14 A0A182HQ46 A0A182UWY8 A0A182TS53 A0A182KSL1 A0A182JBS3 A0A182XNC5 A0A1B0EWS7 A0A1B6G9H8 Q170L4 A0A1Q3FPJ2 A0A1Q3FP78 U5EYT8 A0A0P4W445 A0A023F2I8 A0A2J7PR62 Q7Q082 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A0P4VWU6 A0A0P4W243 A0A224XEU1 V5GQI0 A0A336LL51 A0A1J1IVV5 A0A2P2HWJ3 A0A131YJI2 A0A224Y7S0 A0A147BHW8 A0A1Y1JWI6 A0A1W4U6S0
A0A0L7LTX0 A0A2H1VRZ9 A0A3S2PHU8 A0A212EH32 A0A182MYL4 A0A1B6F720 A0A1L8E3B7 A0A1L8E3B6 A0A1B6JTC9 A0A182Y1R5 A0A182R5C7 A0A182JZY3 A0A182PBS9 A0A2M4CJ21 A0A182W944 A0A182M9X5 A0A182FU59 A0A182QL14 A0A182HQ46 A0A182UWY8 A0A182TS53 A0A182KSL1 A0A182JBS3 A0A182XNC5 A0A1B0EWS7 A0A1B6G9H8 Q170L4 A0A1Q3FPJ2 A0A1Q3FP78 U5EYT8 A0A0P4W445 A0A023F2I8 A0A2J7PR62 Q7Q082 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A0P4VWU6 A0A0P4W243 A0A224XEU1 V5GQI0 A0A336LL51 A0A1J1IVV5 A0A2P2HWJ3 A0A131YJI2 A0A224Y7S0 A0A147BHW8 A0A1Y1JWI6 A0A1W4U6S0
PDB
3Q13
E-value=8.02378e-32,
Score=344
Ontologies
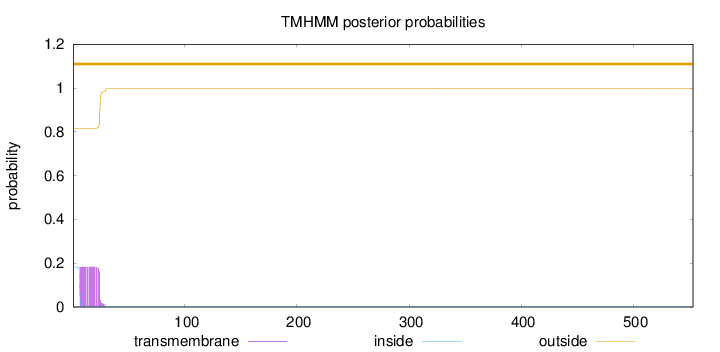
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.977716
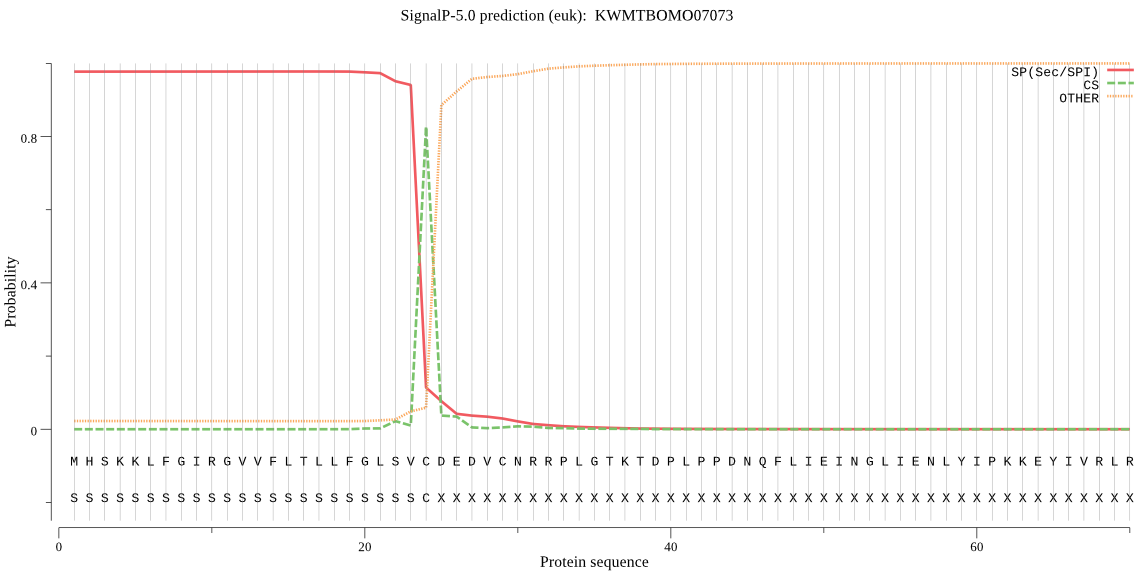
Length:
553
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.38348
Exp number, first 60 AAs:
3.37595
Total prob of N-in:
0.18315
outside
1 - 553
Population Genetic Test Statistics
Pi
207.849585
Theta
156.852676
Tajima's D
0.498639
CLR
0.167939
CSRT
0.512574371281436
Interpretation
Uncertain
