Gene
KWMTBOMO07072
Pre Gene Modal
BGIBMGA010437
Annotation
PREDICTED:_tubulin-specific_chaperone_C_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.834
Sequence
CDS
ATGAATGATAAACATTCTGTCTTAGAACGTCTGAGCCGCAGAGATGCTCAGAGATTAGAAAAACTACAGAAGGCGCATAAGGCAAGAGAAGAAGTAGATGCATGTGATGAAAACGAAGATTACTTTTCGGGTTCATTTAAAATCAAATCTGAATATGTTGAAGATCTTTTAGCTCAAGTACCAACAATAGACAACAGCATGCTCCCATCTCACTTTGACATCATCAAAAAGGAAATTAATGAACTACAGAAATATGTGGTAACATCAACATTTTTCCTTAAGGAATTCAATTTGAGAAAATTTCTTGGCATTGTGCAAAATTTACAAACTAAATGTTATGAATTAGAAGACAGATATGTTCCTCGCAAGAAATTTGGCTTTTCAAAGAAAAAGCTTCCAAAATCTCATAGTCAAAAACAAAATTTAATGGATGAAGCTGATGGAGCAGCTAAAAAAGACAGTAACAAATGGGATGAAAATCTATTTGGATTCAGTTCAAAAGAAAATGAAGTTTTGCTCTTAGAAAATGATGAATTGTACCGAAGAGATGTGGCTTTACGTAATTTGAAAAATTGCACTATAGGTCTCAAAGGTTTAATGGGAACATTACATTTGATCAATTTGGAAAACTGCATAGTTCTTTCTGGGCCGGTAACATCATCAGTATTTGTTGATAAGTGCACAAATTGTAAAATAGTTATTGCATGCCAACAGCTTCGTATGCACACATCAACAAAGTGTGATGTTTATTTGCATGTAACTAGTAAAGGTATTGTAGAGGACTGTTCCCAAATACGCACAGCACCATATAATTTGTTCTATGATAACTTGGAGAAGCATTATGAATCATCCAGTTTAGATAGAAATTCAAATAACTGGGATAATTTGGATGATTTTAATTGGTTAGCTCCTGATGTAAGTTCACCAAATTGGTCGGTTTTAGATGTTTCCGAAAGAATTGATTCATGGGACGATTGGTGGTCTAAGATTCCTTTGTAA
Protein
MNDKHSVLERLSRRDAQRLEKLQKAHKAREEVDACDENEDYFSGSFKIKSEYVEDLLAQVPTIDNSMLPSHFDIIKKEINELQKYVVTSTFFLKEFNLRKFLGIVQNLQTKCYELEDRYVPRKKFGFSKKKLPKSHSQKQNLMDEADGAAKKDSNKWDENLFGFSSKENEVLLLENDELYRRDVALRNLKNCTIGLKGLMGTLHLINLENCIVLSGPVTSSVFVDKCTNCKIVIACQQLRMHTSTKCDVYLHVTSKGIVEDCSQIRTAPYNLFYDNLEKHYESSSLDRNSNNWDNLDDFNWLAPDVSSPNWSVLDVSERIDSWDDWWSKIPL
Summary
Uniprot
H9JLN6
A0A1E1W7X5
A0A2A4K1P7
S4PDP3
A0A194PJM5
A0A212EH44
+ More
A0A194QNH5 A0A0L7LTX0 D2A1K6 A0A067R3R8 A0A0C4URE0 U4UUB7 A0A0T6AYV4 N6TQY4 A0A1Y1K5Q5 A0A1S3HD74 A0A0M8ZRW0 A0A1W4X219 E2BFJ3 E2APQ6 A0A1S3HML1 K7IWH1 A0A088AAY1 A0A232FDG5 A0A2A3EEB1 V9IJK8 A0A026WL76 A0A195E244 A0A195CN74 A0A151JVG9 A0A151WGQ6 V9KXV9 K4GLL9 A0A1L8E572 A0A3B4AJ08 W4Z2R5 A0A2C9JVM6 F4X0X0 H3BDX1 W5NLG1 A0A158N9F7 A0A023EQD5 A0A2T7NYB2 K4FY42 A0A3N0XNN6 A0A2J7PK76 A0A154PFX0 A0A3B3BKM7 A0A1B0CQZ5 A0A182G0P9 A0A1B6DSI0 A0A2U9CN41 A0A151HZ60 C3ZQC0 A0A3B4Z547 H2MX40 I3KY93 A0A3B4F6B1 A0A0E9X5B7 A0A2K6BG00 A0A3P9B8H8 A0A3B3SHQ8 A0A1B0DB62 A0A3Q3BW06 A0A3P8RCL9 A0A1W4ZLW3 G3NFV9 A0A3P8W3C6 A0A3B4ELI1 A0A1L8EIC3 A0A3Q3A6H1 H2ZJ53 A0A1L8EII5 Q0IF54 A0A182X0F0 A0A3Q1HFV9 A0A182VUX3 A0A3Q0RV51 A0A1S4HE66 A0A2K5NU45 A0A182IE64 A0A182RGD3 A0A2K5XV01 G7MPC3 E0VZF4 F1RUS8 F7BNI0 I2CY78 A0A1S4FDV9 A0A369SA37 A0A0L8FZC9 A0A3Q4H0B7 A0A2K5W8E9 A0A096MKM5 B3S3X9 A0A182PA81 A0A2K5IG59 A0A182ULK2 A0A182R1C5 A0A3Q2Z3R6
A0A194QNH5 A0A0L7LTX0 D2A1K6 A0A067R3R8 A0A0C4URE0 U4UUB7 A0A0T6AYV4 N6TQY4 A0A1Y1K5Q5 A0A1S3HD74 A0A0M8ZRW0 A0A1W4X219 E2BFJ3 E2APQ6 A0A1S3HML1 K7IWH1 A0A088AAY1 A0A232FDG5 A0A2A3EEB1 V9IJK8 A0A026WL76 A0A195E244 A0A195CN74 A0A151JVG9 A0A151WGQ6 V9KXV9 K4GLL9 A0A1L8E572 A0A3B4AJ08 W4Z2R5 A0A2C9JVM6 F4X0X0 H3BDX1 W5NLG1 A0A158N9F7 A0A023EQD5 A0A2T7NYB2 K4FY42 A0A3N0XNN6 A0A2J7PK76 A0A154PFX0 A0A3B3BKM7 A0A1B0CQZ5 A0A182G0P9 A0A1B6DSI0 A0A2U9CN41 A0A151HZ60 C3ZQC0 A0A3B4Z547 H2MX40 I3KY93 A0A3B4F6B1 A0A0E9X5B7 A0A2K6BG00 A0A3P9B8H8 A0A3B3SHQ8 A0A1B0DB62 A0A3Q3BW06 A0A3P8RCL9 A0A1W4ZLW3 G3NFV9 A0A3P8W3C6 A0A3B4ELI1 A0A1L8EIC3 A0A3Q3A6H1 H2ZJ53 A0A1L8EII5 Q0IF54 A0A182X0F0 A0A3Q1HFV9 A0A182VUX3 A0A3Q0RV51 A0A1S4HE66 A0A2K5NU45 A0A182IE64 A0A182RGD3 A0A2K5XV01 G7MPC3 E0VZF4 F1RUS8 F7BNI0 I2CY78 A0A1S4FDV9 A0A369SA37 A0A0L8FZC9 A0A3Q4H0B7 A0A2K5W8E9 A0A096MKM5 B3S3X9 A0A182PA81 A0A2K5IG59 A0A182ULK2 A0A182R1C5 A0A3Q2Z3R6
Pubmed
19121390
23622113
26354079
22118469
26227816
18362917
+ More
19820115 24845553 25692689 23537049 28004739 20798317 20075255 28648823 24508170 30249741 24402279 23056606 25463417 15562597 21719571 9215903 21347285 24945155 29451363 26483478 18563158 17554307 25186727 25613341 29240929 24487278 17510324 12364791 22002653 20566863 17431167 25319552 30042472 18719581
19820115 24845553 25692689 23537049 28004739 20798317 20075255 28648823 24508170 30249741 24402279 23056606 25463417 15562597 21719571 9215903 21347285 24945155 29451363 26483478 18563158 17554307 25186727 25613341 29240929 24487278 17510324 12364791 22002653 20566863 17431167 25319552 30042472 18719581
EMBL
BABH01005317
GDQN01008007
GDQN01001868
GDQN01000096
JAT83047.1
JAT89186.1
+ More
JAT90958.1 NWSH01000232 PCG78167.1 GAIX01003586 JAA88974.1 KQ459603 KPI92924.1 AGBW02014975 OWR40791.1 KQ461194 KPJ06909.1 JTDY01000095 KOB78897.1 KQ971338 EFA02681.1 KK852723 KDR17688.1 KJ534556 AIY54297.1 KB632373 ERL93786.1 LJIG01022522 KRT80132.1 APGK01058090 KB741286 ENN70731.1 GEZM01095076 JAV55520.1 KQ435881 KOX69855.1 GL448037 EFN85515.1 GL441573 EFN64582.1 NNAY01000440 OXU28387.1 KZ288267 PBC30067.1 JR050343 AEY61245.1 KK107154 QOIP01000001 EZA56807.1 RLU27165.1 KQ979814 KYN18974.1 KQ977565 KYN01922.1 KQ981713 KYN37380.1 KQ983152 KYQ47020.1 JW871513 AFP04031.1 JX212523 AFM90837.1 GFDF01000268 JAV13816.1 AAGJ04112738 GL888503 EGI59912.1 AFYH01017047 AHAT01001701 ADTU01009631 GAPW01002295 JAC11303.1 PZQS01000008 PVD26168.1 JX052689 AFK10917.1 RJVU01067793 ROI81947.1 NEVH01024940 PNF16736.1 KQ434886 KZC10080.1 AJWK01024246 JXUM01037127 JXUM01037128 JXUM01037129 JXUM01037130 JXUM01037131 GEDC01008664 JAS28634.1 CP026260 AWP17978.1 KQ976712 KYM76974.1 GG666661 EEN45176.1 AERX01030861 GBXM01010808 JAH97769.1 AJVK01004625 AJVK01004626 AJVK01004627 GFDG01000395 JAV18404.1 GFDG01000396 JAV18403.1 CH477394 EAT41884.1 AAAB01008839 APCN01004570 CM001256 EHH18337.1 DS235852 EEB18760.1 AEMK02000055 JSUE03030892 JV667250 AFJ71913.1 NOWV01000035 RDD43649.1 KQ425015 KOF70101.1 AQIA01051831 AHZZ02024664 DS985249 EDV22547.1 AXCN02000858
JAT90958.1 NWSH01000232 PCG78167.1 GAIX01003586 JAA88974.1 KQ459603 KPI92924.1 AGBW02014975 OWR40791.1 KQ461194 KPJ06909.1 JTDY01000095 KOB78897.1 KQ971338 EFA02681.1 KK852723 KDR17688.1 KJ534556 AIY54297.1 KB632373 ERL93786.1 LJIG01022522 KRT80132.1 APGK01058090 KB741286 ENN70731.1 GEZM01095076 JAV55520.1 KQ435881 KOX69855.1 GL448037 EFN85515.1 GL441573 EFN64582.1 NNAY01000440 OXU28387.1 KZ288267 PBC30067.1 JR050343 AEY61245.1 KK107154 QOIP01000001 EZA56807.1 RLU27165.1 KQ979814 KYN18974.1 KQ977565 KYN01922.1 KQ981713 KYN37380.1 KQ983152 KYQ47020.1 JW871513 AFP04031.1 JX212523 AFM90837.1 GFDF01000268 JAV13816.1 AAGJ04112738 GL888503 EGI59912.1 AFYH01017047 AHAT01001701 ADTU01009631 GAPW01002295 JAC11303.1 PZQS01000008 PVD26168.1 JX052689 AFK10917.1 RJVU01067793 ROI81947.1 NEVH01024940 PNF16736.1 KQ434886 KZC10080.1 AJWK01024246 JXUM01037127 JXUM01037128 JXUM01037129 JXUM01037130 JXUM01037131 GEDC01008664 JAS28634.1 CP026260 AWP17978.1 KQ976712 KYM76974.1 GG666661 EEN45176.1 AERX01030861 GBXM01010808 JAH97769.1 AJVK01004625 AJVK01004626 AJVK01004627 GFDG01000395 JAV18404.1 GFDG01000396 JAV18403.1 CH477394 EAT41884.1 AAAB01008839 APCN01004570 CM001256 EHH18337.1 DS235852 EEB18760.1 AEMK02000055 JSUE03030892 JV667250 AFJ71913.1 NOWV01000035 RDD43649.1 KQ425015 KOF70101.1 AQIA01051831 AHZZ02024664 DS985249 EDV22547.1 AXCN02000858
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000007266 UP000027135 UP000030742 UP000019118 UP000085678 UP000053105 UP000192223 UP000008237 UP000000311 UP000002358 UP000005203 UP000215335 UP000242457 UP000053097 UP000279307 UP000078492 UP000078542 UP000078541 UP000075809 UP000261520 UP000007110 UP000076420 UP000007755 UP000008672 UP000018468 UP000005205 UP000245119 UP000235965 UP000076502 UP000261560 UP000092461 UP000069940 UP000246464 UP000078540 UP000001554 UP000261400 UP000001038 UP000005207 UP000261460 UP000233120 UP000265160 UP000261540 UP000092462 UP000264840 UP000265100 UP000192224 UP000007635 UP000265120 UP000261440 UP000264800 UP000007875 UP000008820 UP000076407 UP000265040 UP000075920 UP000261340 UP000233060 UP000075840 UP000075900 UP000233140 UP000009046 UP000008227 UP000006718 UP000253843 UP000053454 UP000261580 UP000233100 UP000028761 UP000009022 UP000075885 UP000233080 UP000075902 UP000075886 UP000264820
UP000007266 UP000027135 UP000030742 UP000019118 UP000085678 UP000053105 UP000192223 UP000008237 UP000000311 UP000002358 UP000005203 UP000215335 UP000242457 UP000053097 UP000279307 UP000078492 UP000078542 UP000078541 UP000075809 UP000261520 UP000007110 UP000076420 UP000007755 UP000008672 UP000018468 UP000005205 UP000245119 UP000235965 UP000076502 UP000261560 UP000092461 UP000069940 UP000246464 UP000078540 UP000001554 UP000261400 UP000001038 UP000005207 UP000261460 UP000233120 UP000265160 UP000261540 UP000092462 UP000264840 UP000265100 UP000192224 UP000007635 UP000265120 UP000261440 UP000264800 UP000007875 UP000008820 UP000076407 UP000265040 UP000075920 UP000261340 UP000233060 UP000075840 UP000075900 UP000233140 UP000009046 UP000008227 UP000006718 UP000253843 UP000053454 UP000261580 UP000233100 UP000028761 UP000009022 UP000075885 UP000233080 UP000075902 UP000075886 UP000264820
Interpro
ProteinModelPortal
H9JLN6
A0A1E1W7X5
A0A2A4K1P7
S4PDP3
A0A194PJM5
A0A212EH44
+ More
A0A194QNH5 A0A0L7LTX0 D2A1K6 A0A067R3R8 A0A0C4URE0 U4UUB7 A0A0T6AYV4 N6TQY4 A0A1Y1K5Q5 A0A1S3HD74 A0A0M8ZRW0 A0A1W4X219 E2BFJ3 E2APQ6 A0A1S3HML1 K7IWH1 A0A088AAY1 A0A232FDG5 A0A2A3EEB1 V9IJK8 A0A026WL76 A0A195E244 A0A195CN74 A0A151JVG9 A0A151WGQ6 V9KXV9 K4GLL9 A0A1L8E572 A0A3B4AJ08 W4Z2R5 A0A2C9JVM6 F4X0X0 H3BDX1 W5NLG1 A0A158N9F7 A0A023EQD5 A0A2T7NYB2 K4FY42 A0A3N0XNN6 A0A2J7PK76 A0A154PFX0 A0A3B3BKM7 A0A1B0CQZ5 A0A182G0P9 A0A1B6DSI0 A0A2U9CN41 A0A151HZ60 C3ZQC0 A0A3B4Z547 H2MX40 I3KY93 A0A3B4F6B1 A0A0E9X5B7 A0A2K6BG00 A0A3P9B8H8 A0A3B3SHQ8 A0A1B0DB62 A0A3Q3BW06 A0A3P8RCL9 A0A1W4ZLW3 G3NFV9 A0A3P8W3C6 A0A3B4ELI1 A0A1L8EIC3 A0A3Q3A6H1 H2ZJ53 A0A1L8EII5 Q0IF54 A0A182X0F0 A0A3Q1HFV9 A0A182VUX3 A0A3Q0RV51 A0A1S4HE66 A0A2K5NU45 A0A182IE64 A0A182RGD3 A0A2K5XV01 G7MPC3 E0VZF4 F1RUS8 F7BNI0 I2CY78 A0A1S4FDV9 A0A369SA37 A0A0L8FZC9 A0A3Q4H0B7 A0A2K5W8E9 A0A096MKM5 B3S3X9 A0A182PA81 A0A2K5IG59 A0A182ULK2 A0A182R1C5 A0A3Q2Z3R6
A0A194QNH5 A0A0L7LTX0 D2A1K6 A0A067R3R8 A0A0C4URE0 U4UUB7 A0A0T6AYV4 N6TQY4 A0A1Y1K5Q5 A0A1S3HD74 A0A0M8ZRW0 A0A1W4X219 E2BFJ3 E2APQ6 A0A1S3HML1 K7IWH1 A0A088AAY1 A0A232FDG5 A0A2A3EEB1 V9IJK8 A0A026WL76 A0A195E244 A0A195CN74 A0A151JVG9 A0A151WGQ6 V9KXV9 K4GLL9 A0A1L8E572 A0A3B4AJ08 W4Z2R5 A0A2C9JVM6 F4X0X0 H3BDX1 W5NLG1 A0A158N9F7 A0A023EQD5 A0A2T7NYB2 K4FY42 A0A3N0XNN6 A0A2J7PK76 A0A154PFX0 A0A3B3BKM7 A0A1B0CQZ5 A0A182G0P9 A0A1B6DSI0 A0A2U9CN41 A0A151HZ60 C3ZQC0 A0A3B4Z547 H2MX40 I3KY93 A0A3B4F6B1 A0A0E9X5B7 A0A2K6BG00 A0A3P9B8H8 A0A3B3SHQ8 A0A1B0DB62 A0A3Q3BW06 A0A3P8RCL9 A0A1W4ZLW3 G3NFV9 A0A3P8W3C6 A0A3B4ELI1 A0A1L8EIC3 A0A3Q3A6H1 H2ZJ53 A0A1L8EII5 Q0IF54 A0A182X0F0 A0A3Q1HFV9 A0A182VUX3 A0A3Q0RV51 A0A1S4HE66 A0A2K5NU45 A0A182IE64 A0A182RGD3 A0A2K5XV01 G7MPC3 E0VZF4 F1RUS8 F7BNI0 I2CY78 A0A1S4FDV9 A0A369SA37 A0A0L8FZC9 A0A3Q4H0B7 A0A2K5W8E9 A0A096MKM5 B3S3X9 A0A182PA81 A0A2K5IG59 A0A182ULK2 A0A182R1C5 A0A3Q2Z3R6
PDB
2YUH
E-value=1.46317e-30,
Score=331
Ontologies
GO
PANTHER
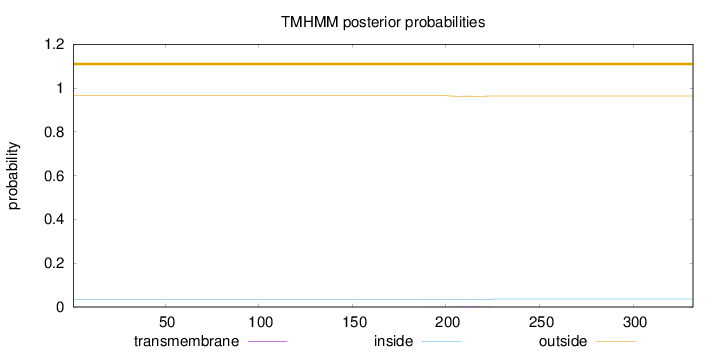
Topology
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06043
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03516
outside
1 - 332
Population Genetic Test Statistics
Pi
249.301412
Theta
237.039891
Tajima's D
0.112613
CLR
0
CSRT
0.4024798760062
Interpretation
Uncertain
