Gene
KWMTBOMO07071 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010523
Annotation
PREDICTED:_N-acetylgalactosaminyltransferase_7_isoform_X2_[Papilio_xuthus]
Full name
Polypeptide N-acetylgalactosaminyltransferase
+ More
N-acetylgalactosaminyltransferase 7
N-acetylgalactosaminyltransferase 7
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
Protein-UDP acetylgalactosaminyltransferase 7
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 7
dGalNAc-T2
Protein-UDP acetylgalactosaminyltransferase 7
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 7
dGalNAc-T2
Location in the cell
Cytoplasmic Reliability : 2.781
Sequence
CDS
ATGTCACAGAAGCATGCCAATGATGTAGCAGAGACTGAGAGTAAATACGGAATGAATATAGCTGCATCAAATGATATTGCAATGAACAGATCGATACCGGATACGCGGCTCGATGAATGCAAATACTGGCACTACCCCGAAGATCTGGCCAAGACATCTGTAATCATCGTCTTCCATAATGAGGGCTTCTCCGTTCTCATGAGAACCGTCCACAGCGTTATTAACAGAACTCCTGCGCAATTTCTGCATGAAGTCGTCTTGGTCGATGATTTCTCCGATAAAGATGACTTAAAAGAAAATTTAGATAATTACATAAAGAGGTGGAATGGTAAAGTGCGTCTTGTGCGTAATGTGCAACGCGAGGGACTGATTCGTACCAGATCGAGGGGGGCCCAAGAAGCGACCGGCGACGTCATAGTCTTCCTTGACGCTCACTGCGAAGTCAATGTCAACTGGCTCCCTCCGCTCTTAGCCCCCATATACAGGGACTACAGAACTATGACCGTGCCCGTTATAGACGGTATAGATTACAACACATTCGAATACAGACCCGTATACCAACACGGAACGAATTACCGCGGTATATTTGAATGGGGTATGTTGTACAAAGAGAACGAAGTGCCCGACAGAGAAGCACATCTTCACAAGCACAAGTCGGAGCCGTACAAGAGTCCGACACACGCTGGAGGCCTGTTCGCCATCAACAGACGATACTTCTTAGAGATAGGCGCGTACGACCCTGGTCTCTTGGTGTGGGGAGGGGAGAACTTTGAGTTGAGCTTCAAAATATGGCAATGCGGTGGCAGCATAGAATGGGTGCCGTGCTCGCGAGTAGGTCACGTATACAGAGCCTTCATGCCGTATACGTTCGGTAACTTGGCCAAAAATCGCAAAGGCTCCCTCATTACGATAAACTACAAACGCGTCATCGAGACCTGGTTCGATGAGGAGCACAAGGAGTACTTCTACACGAGGGAGCCGATGGCGCGCTTCTTGGACATGGGCGACATAAGCGAACAGGTGGCCTTAAAAGAACGCTTGAAGTGCAAAAGCTTCGGTTGGTTCATGGAAAACGTTGCGTACGATGTGTACGATAAGTTCCCGAAGCTCCCTAAGAACGTTCACTGGGGGATGGTGAAGAACAAAGCGACTGGTGCCTGCTTGGATACAATGGGGAAAGCCGCTCCGGCCTATATCGGTACGTCGTCGTGCCACGGGATGGGCAACAGCCAGTTGTTCAGGTTGAATGAGGCCGGCCAGCTCGGCGTCGGCGAGCGGTGCGTGGAGACCGACGGAGATAATGTTAAACAGGCCATTTGCAGACTCGGCACGGTTGACGGACCGTGGAGCTACAACGAGGAGCGCCATCAGCTGGTGCACCGGCTACACGGGCACTGCCTCACGCTGCAACCTCACTCCGGGCAGCTCGGCCTGGCGCCCTGCGACCCAAACAACACATACCAGCAGTGGACCGTCAAACAGAAAACGCCCAACTGACGAGCATCCATAACTAACCTTGACAAAGAGCTGATTCTCGATAAAGATCTGTTTCAAAACTTCACTGGGATGTCCAAAAGTGACTTCGAATATTTGTTGAATACAATTGGACCAAGAATCGAAAAAGGAGATACAAACATGAAAAAGGCAAGGGGAGGTGAGCGACGGAAAAGAGATGGACCTATACATCAGAGCAAAGACGATATACGGCAATATTGCAGTGAAGTCAAAGCGGGATAA
Protein
MSQKHANDVAETESKYGMNIAASNDIAMNRSIPDTRLDECKYWHYPEDLAKTSVIIVFHNEGFSVLMRTVHSVINRTPAQFLHEVVLVDDFSDKDDLKENLDNYIKRWNGKVRLVRNVQREGLIRTRSRGAQEATGDVIVFLDAHCEVNVNWLPPLLAPIYRDYRTMTVPVIDGIDYNTFEYRPVYQHGTNYRGIFEWGMLYKENEVPDREAHLHKHKSEPYKSPTHAGGLFAINRRYFLEIGAYDPGLLVWGGENFELSFKIWQCGGSIEWVPCSRVGHVYRAFMPYTFGNLAKNRKGSLITINYKRVIETWFDEEHKEYFYTREPMARFLDMGDISEQVALKERLKCKSFGWFMENVAYDVYDKFPKLPKNVHWGMVKNKATGACLDTMGKAAPAYIGTSSCHGMGNSQLFRLNEAGQLGVGERCVETDGDNVKQAICRLGTVDGPWSYNEERHQLVHRLHGHCLTLQPHSGQLGLAPCDPNNTYQQWTVKQKTPNXRASITNLDKELILDKDLFQNFTGMSKSDFEYLLNTIGPRIEKGDTNMKKARGGERRKRDGPIHQSKDDIRQYCSEVKAG
Summary
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Keywords
Complete proteome
Disulfide bond
Glycoprotein
Glycosyltransferase
Golgi apparatus
Lectin
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain N-acetylgalactosaminyltransferase 7
Uniprot
L7WM53
H9JLX1
A0A2H1VQD2
A0A194PP64
A0A194QN11
A0A2A4JQI3
+ More
A0A212EH41 A0A3S2NXY9 A0A1J1I0Y3 A0A1J1HZ59 A0A1Y1MRD7 A0A1B6CZF7 A0A1W4WX25 A0A1L8E219 A0A1L8E2H4 D2A249 A0A0L0C0P2 E2B8Q3 U5EM56 A0A0K8USE7 A0A0K8U016 A0A034VVC5 A0A0A1X9D8 T1PIY7 A0A0J7KZQ8 W8BIB6 K7ILV4 A0A154PQ83 E9IGN7 A0A2J7QQP9 Q17CM9 E2AX58 A0A1B6ITM6 A0A1B6H033 A0A1I8P174 B4NEM0 A0A182GZ72 A0A0M8ZUX5 A0A232FK05 A0A3B0KIV4 F4WQT7 A0A1Q3G303 Q29G11 A0A224XKM4 B4Q2Z3 A0A195EG02 A0A195ERQ7 B4I6T4 B4JJ43 B4MD68 A0A1W4UDI3 B3NTE9 X2JEA3 Q8MV48 A0A0N7ZAH1 A0A0P8Y0U8 A0A1B6ITI5 A0A0L7R4V4 B3MWP7 A0A151X3R5 A0A1A9WAR1 A0A195CZN3 A0A0M5J3I3 A0A195BQ00 A0A3L8DKW0 A0A336M2W9 A0A026W9A9 A0A1A9Y010 A0A067R3V9 A0A0C9QYV6 A0A182QEQ6 A0A1B0BYR3 A0A182LUG0 A0A182W5X1 A0A0A9WWW6 A0A182RKP6 A0A182J8H2 A0A1B0GAN4 A0A1A9VD82 A0A182US21 A0A1A9ZP58 A0A182UDT8 A0A182X7F3 Q7QEH0 A0A182I9Y7 A0A182N0U7 A0A088ANK9 A0A084WFL2 U4TWW9 A0A182S5T3 A0A182YH11 T1ID07 A0A182FJ66 A0A2A3EI05 A0A023F529 A0A2M3Z0S9 A0A2M4BHU2 A0A2M4BHW2 W5JDN9 A0A0M4ET62
A0A212EH41 A0A3S2NXY9 A0A1J1I0Y3 A0A1J1HZ59 A0A1Y1MRD7 A0A1B6CZF7 A0A1W4WX25 A0A1L8E219 A0A1L8E2H4 D2A249 A0A0L0C0P2 E2B8Q3 U5EM56 A0A0K8USE7 A0A0K8U016 A0A034VVC5 A0A0A1X9D8 T1PIY7 A0A0J7KZQ8 W8BIB6 K7ILV4 A0A154PQ83 E9IGN7 A0A2J7QQP9 Q17CM9 E2AX58 A0A1B6ITM6 A0A1B6H033 A0A1I8P174 B4NEM0 A0A182GZ72 A0A0M8ZUX5 A0A232FK05 A0A3B0KIV4 F4WQT7 A0A1Q3G303 Q29G11 A0A224XKM4 B4Q2Z3 A0A195EG02 A0A195ERQ7 B4I6T4 B4JJ43 B4MD68 A0A1W4UDI3 B3NTE9 X2JEA3 Q8MV48 A0A0N7ZAH1 A0A0P8Y0U8 A0A1B6ITI5 A0A0L7R4V4 B3MWP7 A0A151X3R5 A0A1A9WAR1 A0A195CZN3 A0A0M5J3I3 A0A195BQ00 A0A3L8DKW0 A0A336M2W9 A0A026W9A9 A0A1A9Y010 A0A067R3V9 A0A0C9QYV6 A0A182QEQ6 A0A1B0BYR3 A0A182LUG0 A0A182W5X1 A0A0A9WWW6 A0A182RKP6 A0A182J8H2 A0A1B0GAN4 A0A1A9VD82 A0A182US21 A0A1A9ZP58 A0A182UDT8 A0A182X7F3 Q7QEH0 A0A182I9Y7 A0A182N0U7 A0A088ANK9 A0A084WFL2 U4TWW9 A0A182S5T3 A0A182YH11 T1ID07 A0A182FJ66 A0A2A3EI05 A0A023F529 A0A2M3Z0S9 A0A2M4BHU2 A0A2M4BHW2 W5JDN9 A0A0M4ET62
EC Number
2.4.1.-
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
26108605 20798317 25348373 25830018 25315136 24495485 20075255 21282665 17510324 17994087 26483478 28648823 21719571 15632085 23185243 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11925450 12829714 12537569 16251381 30249741 24508170 24845553 25401762 26823975 12364791 14747013 17210077 24438588 23537049 25244985 25474469 20920257 23761445
26108605 20798317 25348373 25830018 25315136 24495485 20075255 21282665 17510324 17994087 26483478 28648823 21719571 15632085 23185243 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11925450 12829714 12537569 16251381 30249741 24508170 24845553 25401762 26823975 12364791 14747013 17210077 24438588 23537049 25244985 25474469 20920257 23761445
EMBL
KC020486
AGC81884.1
BABH01005317
ODYU01003798
SOQ43007.1
KQ459603
+ More
KPI92925.1 KQ461194 KPJ06908.1 NWSH01000775 PCG74297.1 AGBW02014975 OWR40790.1 RSAL01000028 RVE51863.1 CVRI01000037 CRK93244.1 CRK93243.1 GEZM01023703 JAV88221.1 GEDC01018635 JAS18663.1 GFDF01001311 JAV12773.1 GFDF01001312 JAV12772.1 KQ971338 EFA02739.1 JRES01001065 KNC25857.1 GL446361 EFN87959.1 GANO01001141 JAB58730.1 GDHF01022697 JAI29617.1 GDHF01032453 JAI19861.1 GAKP01012890 JAC46062.1 GBXI01012583 GBXI01006972 JAD01709.1 JAD07320.1 KA648644 AFP63273.1 LBMM01001595 KMQ96042.1 GAMC01009812 JAB96743.1 KQ435035 KZC14063.1 GL763054 EFZ20244.1 NEVH01012086 PNF30893.1 CH477307 EAT44068.1 GL443520 EFN62008.1 GECU01034289 GECU01017415 JAS73417.1 JAS90291.1 GECZ01001761 JAS68008.1 CH964239 EDW82189.1 JXUM01098952 JXUM01098953 JXUM01098954 KQ564458 KXJ72248.1 KQ435833 KOX71640.1 NNAY01000091 OXU31071.1 OUUW01000011 SPP86379.1 GL888275 EGI63450.1 GFDL01000884 JAV34161.1 CH379065 EAL31418.1 GFTR01007274 JAW09152.1 CM000162 EDX02727.1 KQ978983 KYN26824.1 KQ981993 KYN30908.1 CH480823 EDW56032.1 CH916370 EDV99595.1 CH940660 EDW58140.1 KRF78143.1 CH954180 EDV46600.1 AE014298 AHN59876.1 AF493067 AY268068 BT016123 AY058660 AY061339 GDRN01099126 JAI58832.1 CH902625 KPU75401.1 GECU01017461 JAS90245.1 KQ414657 KOC65806.1 EDV35032.1 KQ982562 KYQ54919.1 KQ977041 KYN06123.1 CP012528 ALC49998.1 KQ976424 KYM88597.1 QOIP01000007 RLU21011.1 UFQT01000116 SSX20328.1 KK107347 EZA52256.1 KK852777 KDR16767.1 GBYB01000899 GBYB01000900 JAG70666.1 JAG70667.1 AXCN02002175 JXJN01009832 JXJN01022772 AXCM01000747 GBHO01030657 GBRD01003603 GDHC01009836 JAG12947.1 JAG62218.1 JAQ08793.1 AXCP01008170 CCAG010022197 AAAB01008847 EAA06901.4 APCN01001075 ATLV01023372 KE525342 KFB49006.1 KB631581 ERL84483.1 ACPB03022137 KZ288254 PBC30806.1 GBBI01002554 JAC16158.1 GGFM01001369 MBW22120.1 GGFJ01003485 MBW52626.1 GGFJ01003486 MBW52627.1 ADMH02001465 ETN62482.1 CP012526 ALC48047.1
KPI92925.1 KQ461194 KPJ06908.1 NWSH01000775 PCG74297.1 AGBW02014975 OWR40790.1 RSAL01000028 RVE51863.1 CVRI01000037 CRK93244.1 CRK93243.1 GEZM01023703 JAV88221.1 GEDC01018635 JAS18663.1 GFDF01001311 JAV12773.1 GFDF01001312 JAV12772.1 KQ971338 EFA02739.1 JRES01001065 KNC25857.1 GL446361 EFN87959.1 GANO01001141 JAB58730.1 GDHF01022697 JAI29617.1 GDHF01032453 JAI19861.1 GAKP01012890 JAC46062.1 GBXI01012583 GBXI01006972 JAD01709.1 JAD07320.1 KA648644 AFP63273.1 LBMM01001595 KMQ96042.1 GAMC01009812 JAB96743.1 KQ435035 KZC14063.1 GL763054 EFZ20244.1 NEVH01012086 PNF30893.1 CH477307 EAT44068.1 GL443520 EFN62008.1 GECU01034289 GECU01017415 JAS73417.1 JAS90291.1 GECZ01001761 JAS68008.1 CH964239 EDW82189.1 JXUM01098952 JXUM01098953 JXUM01098954 KQ564458 KXJ72248.1 KQ435833 KOX71640.1 NNAY01000091 OXU31071.1 OUUW01000011 SPP86379.1 GL888275 EGI63450.1 GFDL01000884 JAV34161.1 CH379065 EAL31418.1 GFTR01007274 JAW09152.1 CM000162 EDX02727.1 KQ978983 KYN26824.1 KQ981993 KYN30908.1 CH480823 EDW56032.1 CH916370 EDV99595.1 CH940660 EDW58140.1 KRF78143.1 CH954180 EDV46600.1 AE014298 AHN59876.1 AF493067 AY268068 BT016123 AY058660 AY061339 GDRN01099126 JAI58832.1 CH902625 KPU75401.1 GECU01017461 JAS90245.1 KQ414657 KOC65806.1 EDV35032.1 KQ982562 KYQ54919.1 KQ977041 KYN06123.1 CP012528 ALC49998.1 KQ976424 KYM88597.1 QOIP01000007 RLU21011.1 UFQT01000116 SSX20328.1 KK107347 EZA52256.1 KK852777 KDR16767.1 GBYB01000899 GBYB01000900 JAG70666.1 JAG70667.1 AXCN02002175 JXJN01009832 JXJN01022772 AXCM01000747 GBHO01030657 GBRD01003603 GDHC01009836 JAG12947.1 JAG62218.1 JAQ08793.1 AXCP01008170 CCAG010022197 AAAB01008847 EAA06901.4 APCN01001075 ATLV01023372 KE525342 KFB49006.1 KB631581 ERL84483.1 ACPB03022137 KZ288254 PBC30806.1 GBBI01002554 JAC16158.1 GGFM01001369 MBW22120.1 GGFJ01003485 MBW52626.1 GGFJ01003486 MBW52627.1 ADMH02001465 ETN62482.1 CP012526 ALC48047.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000283053
+ More
UP000183832 UP000192223 UP000007266 UP000037069 UP000008237 UP000095301 UP000036403 UP000002358 UP000076502 UP000235965 UP000008820 UP000000311 UP000095300 UP000007798 UP000069940 UP000249989 UP000053105 UP000215335 UP000268350 UP000007755 UP000001819 UP000002282 UP000078492 UP000078541 UP000001292 UP000001070 UP000008792 UP000192221 UP000008711 UP000000803 UP000007801 UP000053825 UP000075809 UP000091820 UP000078542 UP000092553 UP000078540 UP000279307 UP000053097 UP000092443 UP000027135 UP000075886 UP000092460 UP000075883 UP000075920 UP000075900 UP000075880 UP000092444 UP000078200 UP000075903 UP000092445 UP000075902 UP000076407 UP000007062 UP000075840 UP000075884 UP000005203 UP000030765 UP000030742 UP000075901 UP000076408 UP000015103 UP000069272 UP000242457 UP000000673
UP000183832 UP000192223 UP000007266 UP000037069 UP000008237 UP000095301 UP000036403 UP000002358 UP000076502 UP000235965 UP000008820 UP000000311 UP000095300 UP000007798 UP000069940 UP000249989 UP000053105 UP000215335 UP000268350 UP000007755 UP000001819 UP000002282 UP000078492 UP000078541 UP000001292 UP000001070 UP000008792 UP000192221 UP000008711 UP000000803 UP000007801 UP000053825 UP000075809 UP000091820 UP000078542 UP000092553 UP000078540 UP000279307 UP000053097 UP000092443 UP000027135 UP000075886 UP000092460 UP000075883 UP000075920 UP000075900 UP000075880 UP000092444 UP000078200 UP000075903 UP000092445 UP000075902 UP000076407 UP000007062 UP000075840 UP000075884 UP000005203 UP000030765 UP000030742 UP000075901 UP000076408 UP000015103 UP000069272 UP000242457 UP000000673
Pfam
Interpro
IPR001173
Glyco_trans_2-like
+ More
IPR029044 Nucleotide-diphossugar_trans
IPR035992 Ricin_B-like_lectins
IPR000772 Ricin_B_lectin
IPR004841 AA-permease/SLC12A_dom
IPR018491 SLC12_C
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR029044 Nucleotide-diphossugar_trans
IPR035992 Ricin_B-like_lectins
IPR000772 Ricin_B_lectin
IPR004841 AA-permease/SLC12A_dom
IPR018491 SLC12_C
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
Gene 3D
CDD
ProteinModelPortal
L7WM53
H9JLX1
A0A2H1VQD2
A0A194PP64
A0A194QN11
A0A2A4JQI3
+ More
A0A212EH41 A0A3S2NXY9 A0A1J1I0Y3 A0A1J1HZ59 A0A1Y1MRD7 A0A1B6CZF7 A0A1W4WX25 A0A1L8E219 A0A1L8E2H4 D2A249 A0A0L0C0P2 E2B8Q3 U5EM56 A0A0K8USE7 A0A0K8U016 A0A034VVC5 A0A0A1X9D8 T1PIY7 A0A0J7KZQ8 W8BIB6 K7ILV4 A0A154PQ83 E9IGN7 A0A2J7QQP9 Q17CM9 E2AX58 A0A1B6ITM6 A0A1B6H033 A0A1I8P174 B4NEM0 A0A182GZ72 A0A0M8ZUX5 A0A232FK05 A0A3B0KIV4 F4WQT7 A0A1Q3G303 Q29G11 A0A224XKM4 B4Q2Z3 A0A195EG02 A0A195ERQ7 B4I6T4 B4JJ43 B4MD68 A0A1W4UDI3 B3NTE9 X2JEA3 Q8MV48 A0A0N7ZAH1 A0A0P8Y0U8 A0A1B6ITI5 A0A0L7R4V4 B3MWP7 A0A151X3R5 A0A1A9WAR1 A0A195CZN3 A0A0M5J3I3 A0A195BQ00 A0A3L8DKW0 A0A336M2W9 A0A026W9A9 A0A1A9Y010 A0A067R3V9 A0A0C9QYV6 A0A182QEQ6 A0A1B0BYR3 A0A182LUG0 A0A182W5X1 A0A0A9WWW6 A0A182RKP6 A0A182J8H2 A0A1B0GAN4 A0A1A9VD82 A0A182US21 A0A1A9ZP58 A0A182UDT8 A0A182X7F3 Q7QEH0 A0A182I9Y7 A0A182N0U7 A0A088ANK9 A0A084WFL2 U4TWW9 A0A182S5T3 A0A182YH11 T1ID07 A0A182FJ66 A0A2A3EI05 A0A023F529 A0A2M3Z0S9 A0A2M4BHU2 A0A2M4BHW2 W5JDN9 A0A0M4ET62
A0A212EH41 A0A3S2NXY9 A0A1J1I0Y3 A0A1J1HZ59 A0A1Y1MRD7 A0A1B6CZF7 A0A1W4WX25 A0A1L8E219 A0A1L8E2H4 D2A249 A0A0L0C0P2 E2B8Q3 U5EM56 A0A0K8USE7 A0A0K8U016 A0A034VVC5 A0A0A1X9D8 T1PIY7 A0A0J7KZQ8 W8BIB6 K7ILV4 A0A154PQ83 E9IGN7 A0A2J7QQP9 Q17CM9 E2AX58 A0A1B6ITM6 A0A1B6H033 A0A1I8P174 B4NEM0 A0A182GZ72 A0A0M8ZUX5 A0A232FK05 A0A3B0KIV4 F4WQT7 A0A1Q3G303 Q29G11 A0A224XKM4 B4Q2Z3 A0A195EG02 A0A195ERQ7 B4I6T4 B4JJ43 B4MD68 A0A1W4UDI3 B3NTE9 X2JEA3 Q8MV48 A0A0N7ZAH1 A0A0P8Y0U8 A0A1B6ITI5 A0A0L7R4V4 B3MWP7 A0A151X3R5 A0A1A9WAR1 A0A195CZN3 A0A0M5J3I3 A0A195BQ00 A0A3L8DKW0 A0A336M2W9 A0A026W9A9 A0A1A9Y010 A0A067R3V9 A0A0C9QYV6 A0A182QEQ6 A0A1B0BYR3 A0A182LUG0 A0A182W5X1 A0A0A9WWW6 A0A182RKP6 A0A182J8H2 A0A1B0GAN4 A0A1A9VD82 A0A182US21 A0A1A9ZP58 A0A182UDT8 A0A182X7F3 Q7QEH0 A0A182I9Y7 A0A182N0U7 A0A088ANK9 A0A084WFL2 U4TWW9 A0A182S5T3 A0A182YH11 T1ID07 A0A182FJ66 A0A2A3EI05 A0A023F529 A0A2M3Z0S9 A0A2M4BHU2 A0A2M4BHW2 W5JDN9 A0A0M4ET62
PDB
6IWR
E-value=2.25635e-134,
Score=1228
Ontologies
PATHWAY
GO
Topology
Subcellular location
Golgi apparatus membrane
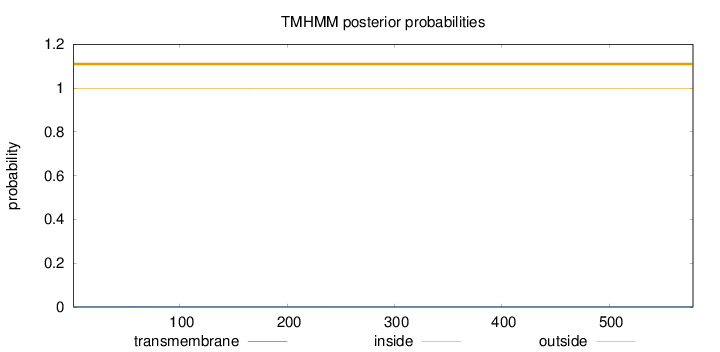
Length:
578
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01431
Exp number, first 60 AAs:
0.00495
Total prob of N-in:
0.00350
outside
1 - 578
Population Genetic Test Statistics
Pi
164.552303
Theta
187.989894
Tajima's D
-0.519777
CLR
0.542996
CSRT
0.233438328083596
Interpretation
Uncertain
