Gene
KWMTBOMO07068 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010439
Annotation
lysozyme_[Manduca_sexta]
Full name
Lysozyme
+ More
Lysozyme c-1
Lysozyme c-1
Alternative Name
1,4-beta-N-acetylmuramidase
Location in the cell
Extracellular Reliability : 4.242
Sequence
CDS
ATGCAGAAGTTAATTATTTTCGCTTTGGTTGTCCTCTGCGTTGGTTCTGAAGCCAAAACGTTCACGAGATGCGGTCTCGTGCATGAGCTGAGGAAACATGGCTTCGAAGAAAATCTCATGAGGAACTGGGTATGTCTGGTCGAGCACGAGAGCAGCCGTGACACATCCAAGACGAACACGAACCGTAACGGCTCGAAGGACTACGGATTGTTCCAGATCAACGACCGGTACTGGTGCAGCAAAGGCGCCAGTCCAGGCAAAGACTGCAACGTTAAGTGCTCCGACCTCCTGACTGACGACATCACTAAGGCAGCGAAATGCGCTAAGAAAATTTACAAACGTCACCGCTTCGATGCCTGGTACGGTTGGAAGAACCACTGCCAGGGCTCTCTGCCTGATATTAGCAGCTGCTAA
Protein
MQKLIIFALVVLCVGSEAKTFTRCGLVHELRKHGFEENLMRNWVCLVEHESSRDTSKTNTNRNGSKDYGLFQINDRYWCSKGASPGKDCNVKCSDLLTDDITKAAKCAKKIYKRHRFDAWYGWKNHCQGSLPDISSC
Summary
Description
Lysozymes have primarily a bacteriolytic function; those in tissues and body fluids are associated with the monocyte-macrophage system and enhance the activity of immunoagents. Active against E.coli and M.luteus.
Lysozymes have primarily a bacteriolytic function; those in tissues and body fluids are associated with the monocyte-macrophage system and enhance the activity of immunoagents.
Lysozymes have primarily a bacteriolytic function; those in tissues and body fluids are associated with the monocyte-macrophage system and enhance the activity of immunoagents.
Catalytic Activity
Hydrolysis of (1->4)-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in a peptidoglycan and between N-acetyl-D-glucosamine residues in chitodextrins.
Subunit
Monomer.
Similarity
Belongs to the glycosyl hydrolase 22 family.
Keywords
3D-structure
Antibiotic
Antimicrobial
Bacteriolytic enzyme
Complete proteome
Direct protein sequencing
Disulfide bond
Glycosidase
Hydrolase
Reference proteome
Signal
Polymorphism
Feature
chain Lysozyme
Uniprot
P48816
Q26363
Q8I808
P50718
Q6QMF0
A0A2H1VQA9
+ More
Q86FK1 B6DZS0 A0A0G3DRZ6 A0A0G3DJB1 A0A1C9CJ68 Q2L7D2 P05105 A0A0G3DPZ6 Q68KS6 A0A2A4JR56 O96862 Q6QEJ9 Q7SID7 Q9GNL4 A0A194QN18 A0A194PHN2 D8L124 F8RGS8 B8YI23 A0A0L7LQW1 P50717 A0A0A7DPR2 A0A3Q8RVM8 A0A3S2TLR1 A9P5Q4 E7DN60 A0A178F6L2 A0A3S2P441 A0A194QNH0 A0A194PP71 S4P2Y8 Q19R28 T2C8V4 A0A3S2LKK5 A0A194PHN8 A0A194PIQ1 A0A346RAE8 P82174 A0A0G3DN23 A0A3S2NN33 A0A3Q8RVZ5 A0A212ETA1 A0A2R7WKI5 T1DGW5 A0A2A4JQM6 A0A2H1VQD1 A0A3Q8RVV0 A0A346RAF0 R4WCI1 B0WEH8 Q9GRF3 B8RJ76 A0A182H4Y7 A0A023EGT6 Q8T3T7 A0A1I8JV61 A0A182VG89 A0A023EFF3 A0A1Y9J0F1 A0A1Y9GKS2 Q17005 A0A182KR20 A0A2A4JPT3 A9XXB6 Q672J4 A0A2H1W700 F5GTS1 F5GTR9 B5M0V6 F5GTS0 F5GTR8 F5GTS2 Q86M91 A0A0G3DJN7 F5GTS3 T2C7G6 I1XB30 D1FPY3 A0A2K8JM74 D1FPX6 A0A023F6U5 A0A240PP26 A0A182MSU2 A0A240PMX4 T1E1T1 A0A182H6E4 R4FPH4 T1D598 A0A067R3S2 A0A240PKQ8 I1XB31 A0A224XNP5 T1IGM3 A0A2Y9D1G9
Q86FK1 B6DZS0 A0A0G3DRZ6 A0A0G3DJB1 A0A1C9CJ68 Q2L7D2 P05105 A0A0G3DPZ6 Q68KS6 A0A2A4JR56 O96862 Q6QEJ9 Q7SID7 Q9GNL4 A0A194QN18 A0A194PHN2 D8L124 F8RGS8 B8YI23 A0A0L7LQW1 P50717 A0A0A7DPR2 A0A3Q8RVM8 A0A3S2TLR1 A9P5Q4 E7DN60 A0A178F6L2 A0A3S2P441 A0A194QNH0 A0A194PP71 S4P2Y8 Q19R28 T2C8V4 A0A3S2LKK5 A0A194PHN8 A0A194PIQ1 A0A346RAE8 P82174 A0A0G3DN23 A0A3S2NN33 A0A3Q8RVZ5 A0A212ETA1 A0A2R7WKI5 T1DGW5 A0A2A4JQM6 A0A2H1VQD1 A0A3Q8RVV0 A0A346RAF0 R4WCI1 B0WEH8 Q9GRF3 B8RJ76 A0A182H4Y7 A0A023EGT6 Q8T3T7 A0A1I8JV61 A0A182VG89 A0A023EFF3 A0A1Y9J0F1 A0A1Y9GKS2 Q17005 A0A182KR20 A0A2A4JPT3 A9XXB6 Q672J4 A0A2H1W700 F5GTS1 F5GTR9 B5M0V6 F5GTS0 F5GTR8 F5GTS2 Q86M91 A0A0G3DJN7 F5GTS3 T2C7G6 I1XB30 D1FPY3 A0A2K8JM74 D1FPX6 A0A023F6U5 A0A240PP26 A0A182MSU2 A0A240PMX4 T1E1T1 A0A182H6E4 R4FPH4 T1D598 A0A067R3S2 A0A240PKQ8 I1XB31 A0A224XNP5 T1IGM3 A0A2Y9D1G9
EC Number
3.2.1.17
Pubmed
7665079
7876591
7517269
8882660
16291089
12831771
+ More
25993013 27575006 2007608 16453632 3840100 9662476 11522783 11290453 26354079 15724965 26227816 21167833 23622113 18615707 24155672 29910977 22118469 24330624 23691247 11122464 17510324 26483478 24945155 12477497 8890741 16137842 12364791 20966253 18154650 15663778 22182526 19166301 12826099 25244985 25474469 24845553
25993013 27575006 2007608 16453632 3840100 9662476 11522783 11290453 26354079 15724965 26227816 21167833 23622113 18615707 24155672 29910977 22118469 24330624 23691247 11122464 17510324 26483478 24945155 12477497 8890741 16137842 12364791 20966253 18154650 15663778 22182526 19166301 12826099 25244985 25474469 24845553
EMBL
L37416
S71028
AAB31190.2
AY164482
AAN87265.1
U38782
+ More
AY533676 AAS48094.1 ODYU01003798 SOQ43009.1 AY251271 AAP03061.1 FJ188380 ACI16106.1 KP056536 AKJ54518.1 KP056538 AKJ54520.1 KX385907 AOM73731.1 DQ353869 ABC73705.1 M60914 X02765 M34923 KP056537 AKJ54519.1 AY684242 AAT94286.1 NWSH01000775 PCG74299.1 U50551 AAD00078.1 AY544998 AAS50157.1 AB048258 BAB20806.1 KQ461194 KPJ06907.1 KQ459603 KPI92926.1 FJ438391 ACR82289.1 HM588761 AEB77713.1 FJ535250 ACL51928.1 JTDY01000298 KOB77820.1 U23786 KF960049 AIW49876.1 MG692502 AZJ51074.1 RSAL01000063 RVE49532.1 EF120625 ABN54797.1 HQ260967 ADU33188.1 LHPN01000102 OAL68001.1 RSAL01000028 RVE51866.1 KPJ06904.1 KPI92930.1 GAIX01008286 JAA84274.1 DQ493869 ABF51015.1 KF146181 AGV28585.1 RSAL01000079 RVE48686.1 KPI92931.1 KPI92928.1 MH200938 AXS59148.1 KP056539 AKJ54521.1 RVE51865.1 MG692501 AZJ51073.1 AGBW02012613 OWR44725.1 KK854991 PTY20156.1 GALA01000032 JAA94820.1 PCG74301.1 SOQ43008.1 MG692500 AZJ51072.1 MH200940 AXS59150.1 AK416888 BAN20103.1 DS231909 EDS45638.1 AJ290428 CH477280 CAC19819.1 EAT44944.1 EZ000501 ACJ64375.1 JXUM01025836 KQ560735 KXJ81084.1 GAPW01005552 JAC08046.1 AY089957 AAM11885.1 GAPW01005551 JAC08047.1 APCN01000195 U28809 DQ007317 AAAB01008807 NWSH01000938 PCG73453.1 EU016395 ABV68862.1 AY693973 AAU09087.1 EAT44943.1 ODYU01006743 SOQ48875.1 JI626234 AEB96469.1 JI626232 AEB96467.1 EU930292 ACH56920.1 JI626233 AEB96468.1 JI626231 AEB96466.1 JI626235 AEB96470.1 AY226458 AB104688 AAO74844.1 BAC82382.1 KP056540 AKJ54522.1 JI626236 AEB96471.1 KF146180 AGV28584.1 JQ754175 AFI81523.1 EZ419883 ACZ28238.1 KY031158 ATU82909.1 EZ419876 ACZ28231.1 GBBI01001606 JAC17106.1 AXCM01004053 GALA01001384 JAA93468.1 JXUM01027018 KQ560774 KXJ80942.1 GAHY01000879 JAA76631.1 GALA01000628 JAA94224.1 KK852779 KDR16709.1 JQ754176 AFI81524.1 GFTR01002321 JAW14105.1 ACPB03012359
AY533676 AAS48094.1 ODYU01003798 SOQ43009.1 AY251271 AAP03061.1 FJ188380 ACI16106.1 KP056536 AKJ54518.1 KP056538 AKJ54520.1 KX385907 AOM73731.1 DQ353869 ABC73705.1 M60914 X02765 M34923 KP056537 AKJ54519.1 AY684242 AAT94286.1 NWSH01000775 PCG74299.1 U50551 AAD00078.1 AY544998 AAS50157.1 AB048258 BAB20806.1 KQ461194 KPJ06907.1 KQ459603 KPI92926.1 FJ438391 ACR82289.1 HM588761 AEB77713.1 FJ535250 ACL51928.1 JTDY01000298 KOB77820.1 U23786 KF960049 AIW49876.1 MG692502 AZJ51074.1 RSAL01000063 RVE49532.1 EF120625 ABN54797.1 HQ260967 ADU33188.1 LHPN01000102 OAL68001.1 RSAL01000028 RVE51866.1 KPJ06904.1 KPI92930.1 GAIX01008286 JAA84274.1 DQ493869 ABF51015.1 KF146181 AGV28585.1 RSAL01000079 RVE48686.1 KPI92931.1 KPI92928.1 MH200938 AXS59148.1 KP056539 AKJ54521.1 RVE51865.1 MG692501 AZJ51073.1 AGBW02012613 OWR44725.1 KK854991 PTY20156.1 GALA01000032 JAA94820.1 PCG74301.1 SOQ43008.1 MG692500 AZJ51072.1 MH200940 AXS59150.1 AK416888 BAN20103.1 DS231909 EDS45638.1 AJ290428 CH477280 CAC19819.1 EAT44944.1 EZ000501 ACJ64375.1 JXUM01025836 KQ560735 KXJ81084.1 GAPW01005552 JAC08046.1 AY089957 AAM11885.1 GAPW01005551 JAC08047.1 APCN01000195 U28809 DQ007317 AAAB01008807 NWSH01000938 PCG73453.1 EU016395 ABV68862.1 AY693973 AAU09087.1 EAT44943.1 ODYU01006743 SOQ48875.1 JI626234 AEB96469.1 JI626232 AEB96467.1 EU930292 ACH56920.1 JI626233 AEB96468.1 JI626231 AEB96466.1 JI626235 AEB96470.1 AY226458 AB104688 AAO74844.1 BAC82382.1 KP056540 AKJ54522.1 JI626236 AEB96471.1 KF146180 AGV28584.1 JQ754175 AFI81523.1 EZ419883 ACZ28238.1 KY031158 ATU82909.1 EZ419876 ACZ28231.1 GBBI01001606 JAC17106.1 AXCM01004053 GALA01001384 JAA93468.1 JXUM01027018 KQ560774 KXJ80942.1 GAHY01000879 JAA76631.1 GALA01000628 JAA94224.1 KK852779 KDR16709.1 JQ754176 AFI81524.1 GFTR01002321 JAW14105.1 ACPB03012359
Proteomes
Pfam
PF00062 Lys
Interpro
SUPFAM
SSF53955
SSF53955
CDD
ProteinModelPortal
P48816
Q26363
Q8I808
P50718
Q6QMF0
A0A2H1VQA9
+ More
Q86FK1 B6DZS0 A0A0G3DRZ6 A0A0G3DJB1 A0A1C9CJ68 Q2L7D2 P05105 A0A0G3DPZ6 Q68KS6 A0A2A4JR56 O96862 Q6QEJ9 Q7SID7 Q9GNL4 A0A194QN18 A0A194PHN2 D8L124 F8RGS8 B8YI23 A0A0L7LQW1 P50717 A0A0A7DPR2 A0A3Q8RVM8 A0A3S2TLR1 A9P5Q4 E7DN60 A0A178F6L2 A0A3S2P441 A0A194QNH0 A0A194PP71 S4P2Y8 Q19R28 T2C8V4 A0A3S2LKK5 A0A194PHN8 A0A194PIQ1 A0A346RAE8 P82174 A0A0G3DN23 A0A3S2NN33 A0A3Q8RVZ5 A0A212ETA1 A0A2R7WKI5 T1DGW5 A0A2A4JQM6 A0A2H1VQD1 A0A3Q8RVV0 A0A346RAF0 R4WCI1 B0WEH8 Q9GRF3 B8RJ76 A0A182H4Y7 A0A023EGT6 Q8T3T7 A0A1I8JV61 A0A182VG89 A0A023EFF3 A0A1Y9J0F1 A0A1Y9GKS2 Q17005 A0A182KR20 A0A2A4JPT3 A9XXB6 Q672J4 A0A2H1W700 F5GTS1 F5GTR9 B5M0V6 F5GTS0 F5GTR8 F5GTS2 Q86M91 A0A0G3DJN7 F5GTS3 T2C7G6 I1XB30 D1FPY3 A0A2K8JM74 D1FPX6 A0A023F6U5 A0A240PP26 A0A182MSU2 A0A240PMX4 T1E1T1 A0A182H6E4 R4FPH4 T1D598 A0A067R3S2 A0A240PKQ8 I1XB31 A0A224XNP5 T1IGM3 A0A2Y9D1G9
Q86FK1 B6DZS0 A0A0G3DRZ6 A0A0G3DJB1 A0A1C9CJ68 Q2L7D2 P05105 A0A0G3DPZ6 Q68KS6 A0A2A4JR56 O96862 Q6QEJ9 Q7SID7 Q9GNL4 A0A194QN18 A0A194PHN2 D8L124 F8RGS8 B8YI23 A0A0L7LQW1 P50717 A0A0A7DPR2 A0A3Q8RVM8 A0A3S2TLR1 A9P5Q4 E7DN60 A0A178F6L2 A0A3S2P441 A0A194QNH0 A0A194PP71 S4P2Y8 Q19R28 T2C8V4 A0A3S2LKK5 A0A194PHN8 A0A194PIQ1 A0A346RAE8 P82174 A0A0G3DN23 A0A3S2NN33 A0A3Q8RVZ5 A0A212ETA1 A0A2R7WKI5 T1DGW5 A0A2A4JQM6 A0A2H1VQD1 A0A3Q8RVV0 A0A346RAF0 R4WCI1 B0WEH8 Q9GRF3 B8RJ76 A0A182H4Y7 A0A023EGT6 Q8T3T7 A0A1I8JV61 A0A182VG89 A0A023EFF3 A0A1Y9J0F1 A0A1Y9GKS2 Q17005 A0A182KR20 A0A2A4JPT3 A9XXB6 Q672J4 A0A2H1W700 F5GTS1 F5GTR9 B5M0V6 F5GTS0 F5GTR8 F5GTS2 Q86M91 A0A0G3DJN7 F5GTS3 T2C7G6 I1XB30 D1FPY3 A0A2K8JM74 D1FPX6 A0A023F6U5 A0A240PP26 A0A182MSU2 A0A240PMX4 T1E1T1 A0A182H6E4 R4FPH4 T1D598 A0A067R3S2 A0A240PKQ8 I1XB31 A0A224XNP5 T1IGM3 A0A2Y9D1G9
PDB
2RSC
E-value=1.79692e-63,
Score=608
Ontologies
GO
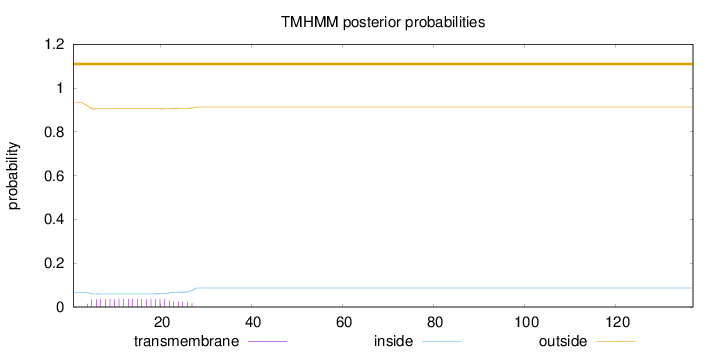
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.998811
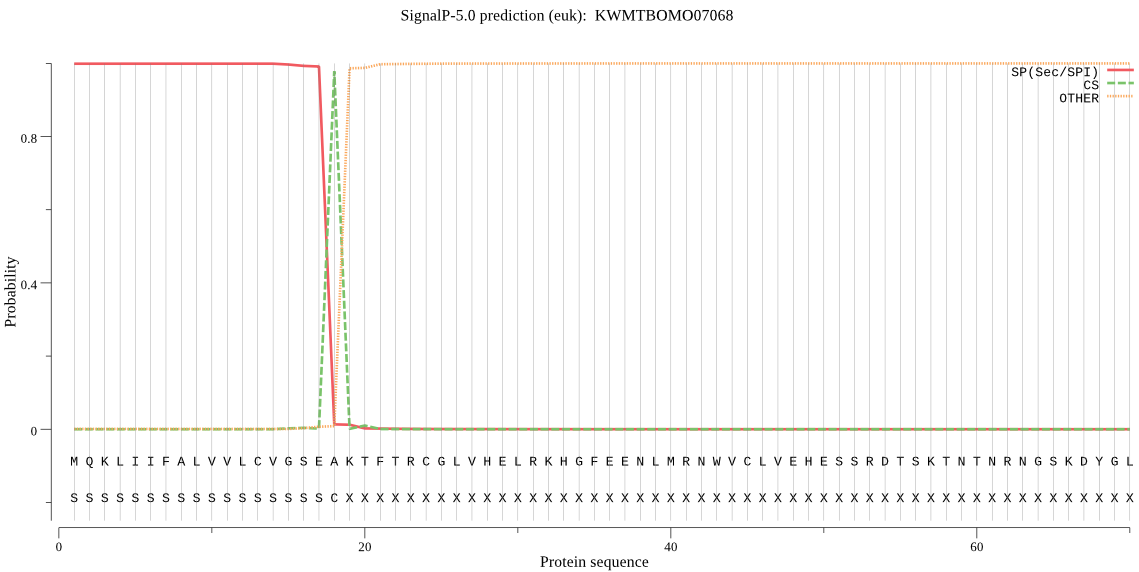
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.76331
Exp number, first 60 AAs:
0.76331
Total prob of N-in:
0.06734
outside
1 - 137
Population Genetic Test Statistics
Pi
206.019109
Theta
154.11265
Tajima's D
0.825442
CLR
0.430645
CSRT
0.612669366531673
Interpretation
Uncertain
