Gene
KWMTBOMO07065
Pre Gene Modal
BGIBMGA010444
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101740376_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.868
Sequence
CDS
ATGCGCGGGAGCACGGCTGGACTCGCTGACACGCTCGCGGGCGGATCCCGAGCCCACGCCGCTTTACTCGCCGCCGCCGACGTCCTCTTCGCAGCAACAGTGGTCGCGCCCGCCGTAGTCGCCTACTGGAAATCAACATGGATCCTTATGGACTTGTATGTGCTACCCGAGCATCCGATCGCTAGTGCGGCCATCAGCGCTGGTTTCGGTCTTCTCTGTGACTTGACGTTTTGCGCGTTACAATCCGAGTTGGCCAAGCTACTGCAGCCAGAGCGCGGTCGGTTGACGTACTACGCGCTGTCGCGTTGTTGCACGTGCGTGGCCGCAGTGGGTTGTGTGGGTGCTTGGCGTGGTGTATGGAACCTGTTGAACGAGTGCACGGGCAACAGCGCGCGGACGCTGCTGTCCACGACGGCGGCGGCGACGCTTTCGCTCGCGGCGCTACGCACATTGCGCAACATCAGCGCGGCGCCATTCGCTGTAGCCGTGGATTCACCGCCCGATTACTTCGCGGTTCCGACAATGTTCCGCACGAACAGTAAAGACATGGCGCTCTACGTGTTGGACTGCGTGTTCTCAGTGAGCGTGGTTGGCTCGCTAGTGGTGTTCGTGTGGCGAGGCTCCTGGGCTCTACTGGACATCTTTATCTTACCAGACAACCTAGTTAAATCTTTGTGGATTTCATTGGCACTAGGTTACGCGCTGGTTATTATGACGTTCTCTCTTCAAGTCCCGATGTGTTGGGCTGCAGCACGTCTCCGAGGCGCGCCGAGACTCCTCCTGGCCGACGTATACCATCTCGTGTCGTTTTTCGCGACGGTCAACGTCTGGCGAGGAGTGTGGGGCTTGCTCAATGTTTATTTCTTCCCTGATTGGCCGAAATTCAGCAACTGGTGCACACATATCGTCAGTTTCTGCCTTCTCGTCTTATTGAACTGTTCCAATTCAATCCTCGTCCGAGGAGTCTACATTGACGCCGAGGAACCGGACGGAGAATGCGTAGTATTCCCGTGTCATTATCTAAGACTATTTGTCCACAAAGAGAGAACTAAGAAGAGACATAAAAGAGCCCTTGCTGCCGCCGCTAAAGAGGGGAGAAGACAAGAAGGCGCTAGTCTACCTCTGCAGCCCGAAGACAAAGTCTAG
Protein
MRGSTAGLADTLAGGSRAHAALLAAADVLFAATVVAPAVVAYWKSTWILMDLYVLPEHPIASAAISAGFGLLCDLTFCALQSELAKLLQPERGRLTYYALSRCCTCVAAVGCVGAWRGVWNLLNECTGNSARTLLSTTAAATLSLAALRTLRNISAAPFAVAVDSPPDYFAVPTMFRTNSKDMALYVLDCVFSVSVVGSLVVFVWRGSWALLDIFILPDNLVKSLWISLALGYALVIMTFSLQVPMCWAAARLRGAPRLLLADVYHLVSFFATVNVWRGVWGLLNVYFFPDWPKFSNWCTHIVSFCLLVLLNCSNSILVRGVYIDAEEPDGECVVFPCHYLRLFVHKERTKKRHKRALAAAAKEGRRQEGASLPLQPEDKV
Summary
Uniprot
A0A2W1BJD1
A0A2A4JEC6
A0A2H1VMB2
A0A194PHP3
A0A212FAA4
A0A0L7KZ67
+ More
A0A1B6LZW8 A0A0T6AZX2 A0A2A3E6N2 E2BXV9 A0A087ZMR9 A0A0L7RC03 F4X588 E2AI68 A0A1J1I465 A0A1L8D9T0 A0A0C9RYM8 A0A151I2W4 A0A151J7H8 A0A195FW03 A0A1B0D2S7 A0A151WV58 A0A146LRE7 A0A151IM72 A0A1Y1NG96 A0A3L8DQQ0 A0A1Q3FS56 A0A146L4R1 A0A336M8G3 A0A154PGG6 A0A139WCA6 K7IQJ1 A0A182J6Q7 J9JTT4 A0A182NTS8 Q7Q4A4 A0A1S4GYM8 A0A2C9GNL2 A0A182TV59 A0A182L0U2 A0A182MCL9 A0A182VAI6 A0A182JY51 A0A182H7R0 A0A084WSE7 Q17L31 U5ESC6 A0A182XL86 A0A2M4CYB5 A0A182Y3E0 A0A1J1I2E8 A0A1S4GXW9 A0A182ICE1 T1HZG5 A0A182PAV5 W5J7T4 A0A182W6M6 A0A182RMC4 A0A182F4Z8 A0A182QD71 B0WQ82 E0VNU7 A0A182G5A6 B4LRB1 B4KI44 B3N4V2 A0A0A1X3P2 A0A1I8P4N0 A0A1W4UID0 B4P070 A0A1I8NAH9 B4Q3K9 Q7K3S3 T1PLB9 T1PE08 A0A0L0BMU6 B3MLL4 B4NLM2 B4I1F6 A0A1B6GG06 B4JPK2 B4G6R2 Q29M09 W8BE27
A0A1B6LZW8 A0A0T6AZX2 A0A2A3E6N2 E2BXV9 A0A087ZMR9 A0A0L7RC03 F4X588 E2AI68 A0A1J1I465 A0A1L8D9T0 A0A0C9RYM8 A0A151I2W4 A0A151J7H8 A0A195FW03 A0A1B0D2S7 A0A151WV58 A0A146LRE7 A0A151IM72 A0A1Y1NG96 A0A3L8DQQ0 A0A1Q3FS56 A0A146L4R1 A0A336M8G3 A0A154PGG6 A0A139WCA6 K7IQJ1 A0A182J6Q7 J9JTT4 A0A182NTS8 Q7Q4A4 A0A1S4GYM8 A0A2C9GNL2 A0A182TV59 A0A182L0U2 A0A182MCL9 A0A182VAI6 A0A182JY51 A0A182H7R0 A0A084WSE7 Q17L31 U5ESC6 A0A182XL86 A0A2M4CYB5 A0A182Y3E0 A0A1J1I2E8 A0A1S4GXW9 A0A182ICE1 T1HZG5 A0A182PAV5 W5J7T4 A0A182W6M6 A0A182RMC4 A0A182F4Z8 A0A182QD71 B0WQ82 E0VNU7 A0A182G5A6 B4LRB1 B4KI44 B3N4V2 A0A0A1X3P2 A0A1I8P4N0 A0A1W4UID0 B4P070 A0A1I8NAH9 B4Q3K9 Q7K3S3 T1PLB9 T1PE08 A0A0L0BMU6 B3MLL4 B4NLM2 B4I1F6 A0A1B6GG06 B4JPK2 B4G6R2 Q29M09 W8BE27
Pubmed
28756777
26354079
22118469
26227816
20798317
21719571
+ More
26823975 28004739 30249741 18362917 19820115 20075255 12364791 20966253 26483478 24438588 17510324 25244985 20920257 23761445 20566863 17994087 18057021 25830018 17550304 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 15632085 24495485
26823975 28004739 30249741 18362917 19820115 20075255 12364791 20966253 26483478 24438588 17510324 25244985 20920257 23761445 20566863 17994087 18057021 25830018 17550304 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 15632085 24495485
EMBL
KZ150015
PZC74978.1
NWSH01001809
PCG70058.1
ODYU01003339
SOQ41969.1
+ More
KQ459603 KPI92936.1 AGBW02009505 OWR50658.1 JTDY01004157 KOB68517.1 GEBQ01010792 JAT29185.1 LJIG01022456 KRT80491.1 KZ288363 PBC26932.1 GL451323 EFN79473.1 KQ414617 KOC68343.1 GL888695 EGI58386.1 GL439655 EFN66867.1 CVRI01000038 CRK94388.1 GFDF01011010 JAV03074.1 GBYB01013076 JAG82843.1 KQ976531 KYM81656.1 KQ979679 KYN19863.1 KQ981215 KYN44623.1 AJVK01002786 KQ982706 KYQ51820.1 GDHC01008235 JAQ10394.1 KQ977063 KYN05980.1 GEZM01003068 JAV96943.1 QOIP01000005 RLU22592.1 GFDL01004660 JAV30385.1 GDHC01015760 JAQ02869.1 UFQT01000603 SSX25651.1 KQ434899 KZC10897.1 KQ971371 KYB25485.1 ABLF02041080 AAAB01008964 EAA12205.3 APCN01000797 AXCM01002004 JXUM01004192 JXUM01004193 KQ560170 KXJ84029.1 ATLV01026496 ATLV01026497 KE525415 KFB53141.1 CH477219 EAT47395.1 GANO01004894 JAB54977.1 GGFL01006023 MBW70201.1 CRK94387.1 ACPB03008637 ADMH02002110 ETN58915.1 AXCN02000803 DS232036 EDS32742.1 DS235354 EEB15053.1 JXUM01043149 KQ561352 KXJ78862.1 CH940649 EDW64581.1 KRF81737.1 KRF81738.1 KRF81739.1 CH933807 EDW12337.2 CH954177 EDV57854.1 GBXI01008338 GBXI01003779 JAD05954.1 JAD10513.1 CM000157 EDW88935.1 CM000361 CM002910 EDX03813.1 KMY88284.1 AE014134 AY058386 AAF52244.1 AAF52245.1 AAL13615.1 AAN10540.1 AFH03563.1 AFH03564.1 AHN54141.1 KA649494 AFP64123.1 KA646183 AFP60812.1 JRES01001631 KNC21332.1 CH902620 EDV31763.1 KPU73612.1 KPU73613.1 CH964274 EDW85261.1 CH480820 EDW54363.1 GECZ01008387 JAS61382.1 CH916372 EDV98832.1 CH479180 EDW29176.1 CH379060 EAL33886.3 GAMC01011304 GAMC01011303 GAMC01011302 JAB95253.1
KQ459603 KPI92936.1 AGBW02009505 OWR50658.1 JTDY01004157 KOB68517.1 GEBQ01010792 JAT29185.1 LJIG01022456 KRT80491.1 KZ288363 PBC26932.1 GL451323 EFN79473.1 KQ414617 KOC68343.1 GL888695 EGI58386.1 GL439655 EFN66867.1 CVRI01000038 CRK94388.1 GFDF01011010 JAV03074.1 GBYB01013076 JAG82843.1 KQ976531 KYM81656.1 KQ979679 KYN19863.1 KQ981215 KYN44623.1 AJVK01002786 KQ982706 KYQ51820.1 GDHC01008235 JAQ10394.1 KQ977063 KYN05980.1 GEZM01003068 JAV96943.1 QOIP01000005 RLU22592.1 GFDL01004660 JAV30385.1 GDHC01015760 JAQ02869.1 UFQT01000603 SSX25651.1 KQ434899 KZC10897.1 KQ971371 KYB25485.1 ABLF02041080 AAAB01008964 EAA12205.3 APCN01000797 AXCM01002004 JXUM01004192 JXUM01004193 KQ560170 KXJ84029.1 ATLV01026496 ATLV01026497 KE525415 KFB53141.1 CH477219 EAT47395.1 GANO01004894 JAB54977.1 GGFL01006023 MBW70201.1 CRK94387.1 ACPB03008637 ADMH02002110 ETN58915.1 AXCN02000803 DS232036 EDS32742.1 DS235354 EEB15053.1 JXUM01043149 KQ561352 KXJ78862.1 CH940649 EDW64581.1 KRF81737.1 KRF81738.1 KRF81739.1 CH933807 EDW12337.2 CH954177 EDV57854.1 GBXI01008338 GBXI01003779 JAD05954.1 JAD10513.1 CM000157 EDW88935.1 CM000361 CM002910 EDX03813.1 KMY88284.1 AE014134 AY058386 AAF52244.1 AAF52245.1 AAL13615.1 AAN10540.1 AFH03563.1 AFH03564.1 AHN54141.1 KA649494 AFP64123.1 KA646183 AFP60812.1 JRES01001631 KNC21332.1 CH902620 EDV31763.1 KPU73612.1 KPU73613.1 CH964274 EDW85261.1 CH480820 EDW54363.1 GECZ01008387 JAS61382.1 CH916372 EDV98832.1 CH479180 EDW29176.1 CH379060 EAL33886.3 GAMC01011304 GAMC01011303 GAMC01011302 JAB95253.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000037510
UP000242457
UP000008237
+ More
UP000005203 UP000053825 UP000007755 UP000000311 UP000183832 UP000078540 UP000078492 UP000078541 UP000092462 UP000075809 UP000078542 UP000279307 UP000076502 UP000007266 UP000002358 UP000075880 UP000007819 UP000075884 UP000007062 UP000075840 UP000075902 UP000075882 UP000075883 UP000075903 UP000075881 UP000069940 UP000249989 UP000030765 UP000008820 UP000076407 UP000076408 UP000015103 UP000075885 UP000000673 UP000075920 UP000075900 UP000069272 UP000075886 UP000002320 UP000009046 UP000008792 UP000009192 UP000008711 UP000095300 UP000192221 UP000002282 UP000095301 UP000000304 UP000000803 UP000037069 UP000007801 UP000007798 UP000001292 UP000001070 UP000008744 UP000001819
UP000005203 UP000053825 UP000007755 UP000000311 UP000183832 UP000078540 UP000078492 UP000078541 UP000092462 UP000075809 UP000078542 UP000279307 UP000076502 UP000007266 UP000002358 UP000075880 UP000007819 UP000075884 UP000007062 UP000075840 UP000075902 UP000075882 UP000075883 UP000075903 UP000075881 UP000069940 UP000249989 UP000030765 UP000008820 UP000076407 UP000076408 UP000015103 UP000075885 UP000000673 UP000075920 UP000075900 UP000069272 UP000075886 UP000002320 UP000009046 UP000008792 UP000009192 UP000008711 UP000095300 UP000192221 UP000002282 UP000095301 UP000000304 UP000000803 UP000037069 UP000007801 UP000007798 UP000001292 UP000001070 UP000008744 UP000001819
PRIDE
Gene 3D
ProteinModelPortal
A0A2W1BJD1
A0A2A4JEC6
A0A2H1VMB2
A0A194PHP3
A0A212FAA4
A0A0L7KZ67
+ More
A0A1B6LZW8 A0A0T6AZX2 A0A2A3E6N2 E2BXV9 A0A087ZMR9 A0A0L7RC03 F4X588 E2AI68 A0A1J1I465 A0A1L8D9T0 A0A0C9RYM8 A0A151I2W4 A0A151J7H8 A0A195FW03 A0A1B0D2S7 A0A151WV58 A0A146LRE7 A0A151IM72 A0A1Y1NG96 A0A3L8DQQ0 A0A1Q3FS56 A0A146L4R1 A0A336M8G3 A0A154PGG6 A0A139WCA6 K7IQJ1 A0A182J6Q7 J9JTT4 A0A182NTS8 Q7Q4A4 A0A1S4GYM8 A0A2C9GNL2 A0A182TV59 A0A182L0U2 A0A182MCL9 A0A182VAI6 A0A182JY51 A0A182H7R0 A0A084WSE7 Q17L31 U5ESC6 A0A182XL86 A0A2M4CYB5 A0A182Y3E0 A0A1J1I2E8 A0A1S4GXW9 A0A182ICE1 T1HZG5 A0A182PAV5 W5J7T4 A0A182W6M6 A0A182RMC4 A0A182F4Z8 A0A182QD71 B0WQ82 E0VNU7 A0A182G5A6 B4LRB1 B4KI44 B3N4V2 A0A0A1X3P2 A0A1I8P4N0 A0A1W4UID0 B4P070 A0A1I8NAH9 B4Q3K9 Q7K3S3 T1PLB9 T1PE08 A0A0L0BMU6 B3MLL4 B4NLM2 B4I1F6 A0A1B6GG06 B4JPK2 B4G6R2 Q29M09 W8BE27
A0A1B6LZW8 A0A0T6AZX2 A0A2A3E6N2 E2BXV9 A0A087ZMR9 A0A0L7RC03 F4X588 E2AI68 A0A1J1I465 A0A1L8D9T0 A0A0C9RYM8 A0A151I2W4 A0A151J7H8 A0A195FW03 A0A1B0D2S7 A0A151WV58 A0A146LRE7 A0A151IM72 A0A1Y1NG96 A0A3L8DQQ0 A0A1Q3FS56 A0A146L4R1 A0A336M8G3 A0A154PGG6 A0A139WCA6 K7IQJ1 A0A182J6Q7 J9JTT4 A0A182NTS8 Q7Q4A4 A0A1S4GYM8 A0A2C9GNL2 A0A182TV59 A0A182L0U2 A0A182MCL9 A0A182VAI6 A0A182JY51 A0A182H7R0 A0A084WSE7 Q17L31 U5ESC6 A0A182XL86 A0A2M4CYB5 A0A182Y3E0 A0A1J1I2E8 A0A1S4GXW9 A0A182ICE1 T1HZG5 A0A182PAV5 W5J7T4 A0A182W6M6 A0A182RMC4 A0A182F4Z8 A0A182QD71 B0WQ82 E0VNU7 A0A182G5A6 B4LRB1 B4KI44 B3N4V2 A0A0A1X3P2 A0A1I8P4N0 A0A1W4UID0 B4P070 A0A1I8NAH9 B4Q3K9 Q7K3S3 T1PLB9 T1PE08 A0A0L0BMU6 B3MLL4 B4NLM2 B4I1F6 A0A1B6GG06 B4JPK2 B4G6R2 Q29M09 W8BE27
Ontologies
GO
PANTHER
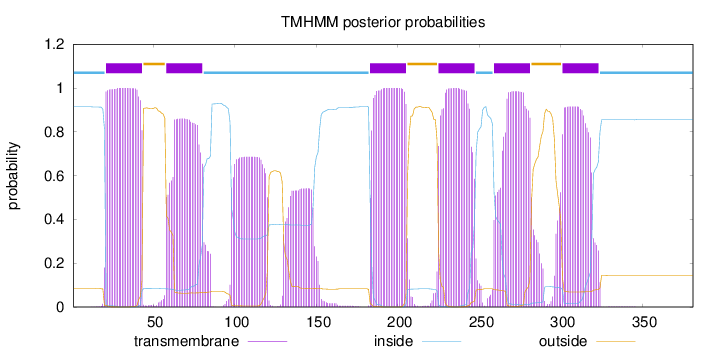
Topology
Length:
381
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
154.33589
Exp number, first 60 AAs:
24.65219
Total prob of N-in:
0.91443
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 57
TMhelix
58 - 80
inside
81 - 182
TMhelix
183 - 205
outside
206 - 224
TMhelix
225 - 247
inside
248 - 258
TMhelix
259 - 281
outside
282 - 300
TMhelix
301 - 323
inside
324 - 381
Population Genetic Test Statistics
Pi
280.052433
Theta
18.749572
Tajima's D
1.4848
CLR
0.357074
CSRT
0.789760511974401
Interpretation
Uncertain
