Gene
KWMTBOMO07063
Pre Gene Modal
BGIBMGA010445
Annotation
PREDICTED:_glucose_1?6-bisphosphate_synthase_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.981
Sequence
CDS
ATGAGTGTGTGTAATAGTGGAGACGTTAAATTAGATGCAGCAGTTAATGCCTGGCTGAAACACGATAAGAATCCCACAACACGTCAAGAAGTATTGGATGATATTAAAAATTCAAGATGGGAGAAATTAAAGAAGACGATGCTACATCGCCAAAAGTTTGGCACGGCTGGTCTACGTGGACGAATGGGTGCTGGATACACGTGTATGAATGATGTTGTGGTTTTACAGACTGCTCAAGGAATATGTTCATATATACAAAAGGTATCCAATAATGCGCAAATAAAAAATGGAATCGTGGTTGGATTTGATGGAAGACATCAGAGTAAAAGATTCGCGGAACTCACAACGAAGGTGTTCATATCTTCATCAATACCGGTCCATTTATTTTCAATGGTTTGTCCGACACCACTCGTGTCGTTTGGCACAATAATGTACGGCGCTGCCGCCGGTGTCATGGTGACGGCCTCCCACAACCCGAAGGAGGACAACGGATACAAAGTTTATTGGTCGAACGGTTCGCAAATTATATCGCCTCATGACGACAACATACTGGATGAGATATTGCAGTGCTTGGACATACCAGACGATCATTGGGATATAACCGAAGTCAGAGCTCACGAATTAGTCCGGGACTGCACAACGGAAGTGACGGATAGGTACATGGCGTACATCCGGGACAGCCTTCACGAGGACGTGCTGGAGACGAACAAGAGAGCCGCCATCGATGTTGTGTACAGCGCGATGCACGGCGTCGGCTACGAATATGTCTTGAAGGCTTTCAAAACGGCCGGATTGAAGGTATCTTACAATATAATATCACTTTACTGGGTGTAG
Protein
MSVCNSGDVKLDAAVNAWLKHDKNPTTRQEVLDDIKNSRWEKLKKTMLHRQKFGTAGLRGRMGAGYTCMNDVVVLQTAQGICSYIQKVSNNAQIKNGIVVGFDGRHQSKRFAELTTKVFISSSIPVHLFSMVCPTPLVSFGTIMYGAAAGVMVTASHNPKEDNGYKVYWSNGSQIISPHDDNILDEILQCLDIPDDHWDITEVRAHELVRDCTTEVTDRYMAYIRDSLHEDVLETNKRAAIDVVYSAMHGVGYEYVLKAFKTAGLKVSYNIISLYWV
Summary
Similarity
Belongs to the phosphohexose mutase family.
Uniprot
H9JLP3
A0A194PJN5
A0A194QN07
A0A3S2TPU5
A0A2A4JE52
A0A2W1BQG6
+ More
A0A212ETF5 A0A2H1VMC8 A0A0L7LMI2 A0A210Q4X0 A0A0N7Z9F3 T1HFV0 V5GV69 F4X483 A0A151II08 A0A1I8PBJ4 A0A023F3Y4 A0A0V0G6E7 A0A1A9W0Z2 A0A0D2WJN3 A0A2J7R8Q8 A0A0L0CKQ6 A0A151JVD8 A0A0K8TLA1 A0A158N9I0 E9IW51 E0VRR4 A0A2P2I8X3 A0A3L8E3D6 A0A232FLN0 A0A195E281 K7IQI9 A0A151I4F8 A0A151XGX6 A0A0J7L3G8 A0A3Q2G2J3 A0A3Q4HKY1 Q17L34 I3JQW5 A0A3P8PWH4 A0A3P9D2X6 A0A3B3W266 B4NN36 A0A026WDR6 A0A146WRM2 A0A087Y0R5 A0A3B3WTW0 A0A336KYE3 A0A0L7RC26 A0A3B3W276 D6X0R3 A0A2C9KCH4 A0A3P9NJZ7 A0A3Q2UVC0 A0A3P9NJZ9 Q58I83 A0A2R5LLS1 M4AE43 A0A336MA45 A0A3B5M426 A0A3B4H8V1 A0A3Q1GCN0 T1P9K7 A0A3B5B3C8 A0A1I8MFS4 D3TP20 A0A3B4BJF8 T2M7M3 A0A3P8TZD1 Q28ZB9 B4GHZ1 A0A0T6B919 B4LPL6 H3CJC9 A0A3Q3IXP9 A0A146N191 A0A088AJC4 A0A3B0JP51 A0A146NQG6 A0A1B0BKV4 A0A3Q1APX9 A0A3R7QKZ5 N6TSP4 A0A0M8ZTG9 U4URF4 B4J7V7 A0A2A3E9E1 A0A1A9WEI5 A0A146WTE9 A0A146LSD4 A0A1A9XY34 B4LPL7 H2RYP0 A0A023EVI7 A0A1Y1N3L1 A0A182H7R3 A0A1A9UU14 A0A0P5XPH8 A0A0P6GCX0 A0A0V0YKC9
A0A212ETF5 A0A2H1VMC8 A0A0L7LMI2 A0A210Q4X0 A0A0N7Z9F3 T1HFV0 V5GV69 F4X483 A0A151II08 A0A1I8PBJ4 A0A023F3Y4 A0A0V0G6E7 A0A1A9W0Z2 A0A0D2WJN3 A0A2J7R8Q8 A0A0L0CKQ6 A0A151JVD8 A0A0K8TLA1 A0A158N9I0 E9IW51 E0VRR4 A0A2P2I8X3 A0A3L8E3D6 A0A232FLN0 A0A195E281 K7IQI9 A0A151I4F8 A0A151XGX6 A0A0J7L3G8 A0A3Q2G2J3 A0A3Q4HKY1 Q17L34 I3JQW5 A0A3P8PWH4 A0A3P9D2X6 A0A3B3W266 B4NN36 A0A026WDR6 A0A146WRM2 A0A087Y0R5 A0A3B3WTW0 A0A336KYE3 A0A0L7RC26 A0A3B3W276 D6X0R3 A0A2C9KCH4 A0A3P9NJZ7 A0A3Q2UVC0 A0A3P9NJZ9 Q58I83 A0A2R5LLS1 M4AE43 A0A336MA45 A0A3B5M426 A0A3B4H8V1 A0A3Q1GCN0 T1P9K7 A0A3B5B3C8 A0A1I8MFS4 D3TP20 A0A3B4BJF8 T2M7M3 A0A3P8TZD1 Q28ZB9 B4GHZ1 A0A0T6B919 B4LPL6 H3CJC9 A0A3Q3IXP9 A0A146N191 A0A088AJC4 A0A3B0JP51 A0A146NQG6 A0A1B0BKV4 A0A3Q1APX9 A0A3R7QKZ5 N6TSP4 A0A0M8ZTG9 U4URF4 B4J7V7 A0A2A3E9E1 A0A1A9WEI5 A0A146WTE9 A0A146LSD4 A0A1A9XY34 B4LPL7 H2RYP0 A0A023EVI7 A0A1Y1N3L1 A0A182H7R3 A0A1A9UU14 A0A0P5XPH8 A0A0P6GCX0 A0A0V0YKC9
Pubmed
19121390
26354079
28756777
22118469
26227816
28812685
+ More
27129103 21719571 25474469 26108605 26369729 21347285 21282665 20566863 30249741 28648823 20075255 25186727 17510324 17994087 24508170 18362917 19820115 15562597 23542700 25315136 20353571 25463417 24065732 15632085 15496914 23537049 26823975 21551351 24945155 28004739 26483478
27129103 21719571 25474469 26108605 26369729 21347285 21282665 20566863 30249741 28648823 20075255 25186727 17510324 17994087 24508170 18362917 19820115 15562597 23542700 25315136 20353571 25463417 24065732 15632085 15496914 23537049 26823975 21551351 24945155 28004739 26483478
EMBL
BABH01005300
KQ459603
KPI92939.1
KQ461194
KPJ06897.1
RSAL01000028
+ More
RVE51871.1 NWSH01001809 PCG70056.1 KZ150015 PZC74980.1 AGBW02012613 OWR44724.1 ODYU01003339 SOQ41967.1 JTDY01000537 KOB76753.1 NEDP02004995 OWF43792.1 GDKW01000707 JAI55888.1 ACPB03018819 GALX01004338 JAB64128.1 GL888628 EGI58751.1 KQ977545 KYN02036.1 GBBI01003048 JAC15664.1 GECL01002547 JAP03577.1 KE346360 KJE89583.1 NEVH01006721 PNF37214.1 JRES01000265 KNC32822.1 KQ981713 KYN37350.1 GDAI01002908 JAI14695.1 ADTU01009654 GL766449 EFZ15238.1 DS235494 EEB16070.1 IACF01004871 LAB70459.1 QOIP01000001 RLU27260.1 NNAY01000048 OXU31565.1 KQ979814 KYN19009.1 KQ976461 KYM84714.1 KQ982169 KYQ59460.1 LBMM01000816 KMQ97422.1 CH477219 EAT47392.1 AERX01027844 CH964282 EDW85775.1 KK107260 EZA54197.1 GCES01053745 JAR32578.1 AYCK01004610 UFQS01000665 UFQT01000665 SSX05960.1 SSX26319.1 KQ414617 KOC68335.1 KQ971371 EFA10132.1 AY944756 AAX47079.1 GGLE01006313 MBY10439.1 UFQS01000603 UFQT01000603 SSX05292.1 SSX25653.1 KA644608 AFP59237.1 CCAG010004670 EZ423172 ADD19448.1 HAAD01001857 CDG68089.1 CM000071 EAL25694.3 CH479183 EDW36111.1 LJIG01009058 KRT83839.1 CH940648 EDW60254.1 GCES01160719 JAQ25603.1 OUUW01000001 SPP74431.1 GCES01152637 JAQ33685.1 JXJN01016040 QCYY01002387 ROT70769.1 APGK01056335 APGK01056336 KB741274 ENN71361.1 KQ435851 KOX70795.1 KB632406 ERL95093.1 CH916367 EDW01163.1 KZ288338 PBC27809.1 GCES01053744 JAR32579.1 GDHC01008460 GDHC01000191 JAQ10169.1 JAQ18438.1 EDW60255.1 GAPW01000562 JAC13036.1 GEZM01013572 GEZM01013571 GEZM01013570 GEZM01013569 JAV92473.1 JXUM01004201 JXUM01004202 KQ560170 KXJ84032.1 GDIP01069188 JAM34527.1 GDIQ01035123 JAN59614.1 JYDU01000007 KRY00557.1
RVE51871.1 NWSH01001809 PCG70056.1 KZ150015 PZC74980.1 AGBW02012613 OWR44724.1 ODYU01003339 SOQ41967.1 JTDY01000537 KOB76753.1 NEDP02004995 OWF43792.1 GDKW01000707 JAI55888.1 ACPB03018819 GALX01004338 JAB64128.1 GL888628 EGI58751.1 KQ977545 KYN02036.1 GBBI01003048 JAC15664.1 GECL01002547 JAP03577.1 KE346360 KJE89583.1 NEVH01006721 PNF37214.1 JRES01000265 KNC32822.1 KQ981713 KYN37350.1 GDAI01002908 JAI14695.1 ADTU01009654 GL766449 EFZ15238.1 DS235494 EEB16070.1 IACF01004871 LAB70459.1 QOIP01000001 RLU27260.1 NNAY01000048 OXU31565.1 KQ979814 KYN19009.1 KQ976461 KYM84714.1 KQ982169 KYQ59460.1 LBMM01000816 KMQ97422.1 CH477219 EAT47392.1 AERX01027844 CH964282 EDW85775.1 KK107260 EZA54197.1 GCES01053745 JAR32578.1 AYCK01004610 UFQS01000665 UFQT01000665 SSX05960.1 SSX26319.1 KQ414617 KOC68335.1 KQ971371 EFA10132.1 AY944756 AAX47079.1 GGLE01006313 MBY10439.1 UFQS01000603 UFQT01000603 SSX05292.1 SSX25653.1 KA644608 AFP59237.1 CCAG010004670 EZ423172 ADD19448.1 HAAD01001857 CDG68089.1 CM000071 EAL25694.3 CH479183 EDW36111.1 LJIG01009058 KRT83839.1 CH940648 EDW60254.1 GCES01160719 JAQ25603.1 OUUW01000001 SPP74431.1 GCES01152637 JAQ33685.1 JXJN01016040 QCYY01002387 ROT70769.1 APGK01056335 APGK01056336 KB741274 ENN71361.1 KQ435851 KOX70795.1 KB632406 ERL95093.1 CH916367 EDW01163.1 KZ288338 PBC27809.1 GCES01053744 JAR32579.1 GDHC01008460 GDHC01000191 JAQ10169.1 JAQ18438.1 EDW60255.1 GAPW01000562 JAC13036.1 GEZM01013572 GEZM01013571 GEZM01013570 GEZM01013569 JAV92473.1 JXUM01004201 JXUM01004202 KQ560170 KXJ84032.1 GDIP01069188 JAM34527.1 GDIQ01035123 JAN59614.1 JYDU01000007 KRY00557.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000218220
UP000007151
+ More
UP000037510 UP000242188 UP000015103 UP000007755 UP000078542 UP000095300 UP000091820 UP000008743 UP000235965 UP000037069 UP000078541 UP000005205 UP000009046 UP000279307 UP000215335 UP000078492 UP000002358 UP000078540 UP000075809 UP000036403 UP000265020 UP000261580 UP000008820 UP000005207 UP000265100 UP000265160 UP000261500 UP000007798 UP000053097 UP000028760 UP000261480 UP000053825 UP000007266 UP000076420 UP000242638 UP000264840 UP000002852 UP000261380 UP000261460 UP000257200 UP000261400 UP000095301 UP000092444 UP000261520 UP000265080 UP000001819 UP000008744 UP000008792 UP000007303 UP000261600 UP000005203 UP000268350 UP000092460 UP000257160 UP000283509 UP000019118 UP000053105 UP000030742 UP000001070 UP000242457 UP000092443 UP000005226 UP000069940 UP000249989 UP000078200 UP000054815
UP000037510 UP000242188 UP000015103 UP000007755 UP000078542 UP000095300 UP000091820 UP000008743 UP000235965 UP000037069 UP000078541 UP000005205 UP000009046 UP000279307 UP000215335 UP000078492 UP000002358 UP000078540 UP000075809 UP000036403 UP000265020 UP000261580 UP000008820 UP000005207 UP000265100 UP000265160 UP000261500 UP000007798 UP000053097 UP000028760 UP000261480 UP000053825 UP000007266 UP000076420 UP000242638 UP000264840 UP000002852 UP000261380 UP000261460 UP000257200 UP000261400 UP000095301 UP000092444 UP000261520 UP000265080 UP000001819 UP000008744 UP000008792 UP000007303 UP000261600 UP000005203 UP000268350 UP000092460 UP000257160 UP000283509 UP000019118 UP000053105 UP000030742 UP000001070 UP000242457 UP000092443 UP000005226 UP000069940 UP000249989 UP000078200 UP000054815
PRIDE
Interpro
ProteinModelPortal
H9JLP3
A0A194PJN5
A0A194QN07
A0A3S2TPU5
A0A2A4JE52
A0A2W1BQG6
+ More
A0A212ETF5 A0A2H1VMC8 A0A0L7LMI2 A0A210Q4X0 A0A0N7Z9F3 T1HFV0 V5GV69 F4X483 A0A151II08 A0A1I8PBJ4 A0A023F3Y4 A0A0V0G6E7 A0A1A9W0Z2 A0A0D2WJN3 A0A2J7R8Q8 A0A0L0CKQ6 A0A151JVD8 A0A0K8TLA1 A0A158N9I0 E9IW51 E0VRR4 A0A2P2I8X3 A0A3L8E3D6 A0A232FLN0 A0A195E281 K7IQI9 A0A151I4F8 A0A151XGX6 A0A0J7L3G8 A0A3Q2G2J3 A0A3Q4HKY1 Q17L34 I3JQW5 A0A3P8PWH4 A0A3P9D2X6 A0A3B3W266 B4NN36 A0A026WDR6 A0A146WRM2 A0A087Y0R5 A0A3B3WTW0 A0A336KYE3 A0A0L7RC26 A0A3B3W276 D6X0R3 A0A2C9KCH4 A0A3P9NJZ7 A0A3Q2UVC0 A0A3P9NJZ9 Q58I83 A0A2R5LLS1 M4AE43 A0A336MA45 A0A3B5M426 A0A3B4H8V1 A0A3Q1GCN0 T1P9K7 A0A3B5B3C8 A0A1I8MFS4 D3TP20 A0A3B4BJF8 T2M7M3 A0A3P8TZD1 Q28ZB9 B4GHZ1 A0A0T6B919 B4LPL6 H3CJC9 A0A3Q3IXP9 A0A146N191 A0A088AJC4 A0A3B0JP51 A0A146NQG6 A0A1B0BKV4 A0A3Q1APX9 A0A3R7QKZ5 N6TSP4 A0A0M8ZTG9 U4URF4 B4J7V7 A0A2A3E9E1 A0A1A9WEI5 A0A146WTE9 A0A146LSD4 A0A1A9XY34 B4LPL7 H2RYP0 A0A023EVI7 A0A1Y1N3L1 A0A182H7R3 A0A1A9UU14 A0A0P5XPH8 A0A0P6GCX0 A0A0V0YKC9
A0A212ETF5 A0A2H1VMC8 A0A0L7LMI2 A0A210Q4X0 A0A0N7Z9F3 T1HFV0 V5GV69 F4X483 A0A151II08 A0A1I8PBJ4 A0A023F3Y4 A0A0V0G6E7 A0A1A9W0Z2 A0A0D2WJN3 A0A2J7R8Q8 A0A0L0CKQ6 A0A151JVD8 A0A0K8TLA1 A0A158N9I0 E9IW51 E0VRR4 A0A2P2I8X3 A0A3L8E3D6 A0A232FLN0 A0A195E281 K7IQI9 A0A151I4F8 A0A151XGX6 A0A0J7L3G8 A0A3Q2G2J3 A0A3Q4HKY1 Q17L34 I3JQW5 A0A3P8PWH4 A0A3P9D2X6 A0A3B3W266 B4NN36 A0A026WDR6 A0A146WRM2 A0A087Y0R5 A0A3B3WTW0 A0A336KYE3 A0A0L7RC26 A0A3B3W276 D6X0R3 A0A2C9KCH4 A0A3P9NJZ7 A0A3Q2UVC0 A0A3P9NJZ9 Q58I83 A0A2R5LLS1 M4AE43 A0A336MA45 A0A3B5M426 A0A3B4H8V1 A0A3Q1GCN0 T1P9K7 A0A3B5B3C8 A0A1I8MFS4 D3TP20 A0A3B4BJF8 T2M7M3 A0A3P8TZD1 Q28ZB9 B4GHZ1 A0A0T6B919 B4LPL6 H3CJC9 A0A3Q3IXP9 A0A146N191 A0A088AJC4 A0A3B0JP51 A0A146NQG6 A0A1B0BKV4 A0A3Q1APX9 A0A3R7QKZ5 N6TSP4 A0A0M8ZTG9 U4URF4 B4J7V7 A0A2A3E9E1 A0A1A9WEI5 A0A146WTE9 A0A146LSD4 A0A1A9XY34 B4LPL7 H2RYP0 A0A023EVI7 A0A1Y1N3L1 A0A182H7R3 A0A1A9UU14 A0A0P5XPH8 A0A0P6GCX0 A0A0V0YKC9
PDB
3I3W
E-value=5.23781e-10,
Score=152
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00030 Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00052 Galactose metabolism - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00500 Starch and sucrose metabolism - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
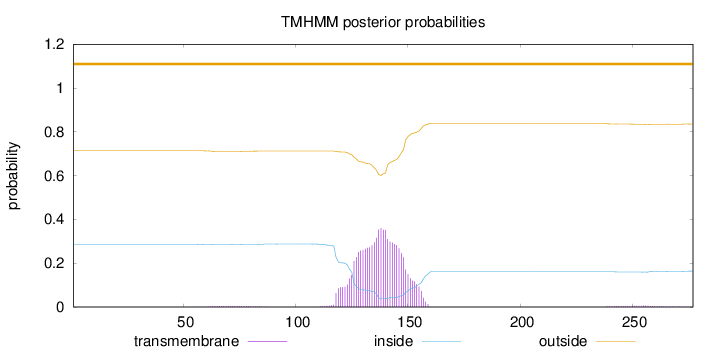
Topology
Length:
277
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.646
Exp number, first 60 AAs:
0.0058
Total prob of N-in:
0.28683
outside
1 - 277
Population Genetic Test Statistics
Pi
251.215743
Theta
163.192545
Tajima's D
1.63534
CLR
0.090978
CSRT
0.818809059547023
Interpretation
Uncertain
