Gene
KWMTBOMO07062
Annotation
Phosphoglucomutase_[Operophtera_brumata]
Full name
Phosphoglucomutase-2
Alternative Name
Glucose phosphomutase 2
Phosphodeoxyribomutase
Phosphopentomutase
Phosphodeoxyribomutase
Phosphopentomutase
Location in the cell
Extracellular Reliability : 1.896
Sequence
CDS
ATGGGCGCACTGCTGGGCTGGTGGCTGCTGACGCAGCACTACAAGAAGGACCCGTCTGCGAATGCCGAGGACCTGTACGTGATCGCCAGCATCGTCAGCTCGAAGATGCTGCGCGCCATCGTCGACGGGAAGGGACAGTTTGTGGAGACACTCACCGGCTTCAAGTGGATGGGTAATTGCTCCCTGCTGCTGGTACAGCAGAACAAGATTCCGGTGTTCGCCTTCGAGGAGGCCCTGGGCTACATGTGCGATGTGCGCGTACCAGACAAAGATGGCGTCTCGGCCGCGGTGCAAGTAAGTATAGTATAG
Protein
MGALLGWWLLTQHYKKDPSANAEDLYVIASIVSSKMLRAIVDGKGQFVETLTGFKWMGNCSLLLVQQNKIPVFAFEEALGYMCDVRVPDKDGVSAAVQVSIV
Summary
Description
Catalyzes the conversion of the nucleoside breakdown products ribose-1-phosphate and deoxyribose-1-phosphate to the corresponding 5-phosphopentoses. May also catalyze the interconversion of glucose-1-phosphate and glucose-6-phosphate. Has low glucose 1,6-bisphosphate synthase activity.
Catalyzes the conversion of the nucleoside breakdown products ribose-1-phosphate and deoxyribose-1-phosphate to the corresponding 5-phosphopentoses. May also catalyze the interconversion of glucose-1-phosphate and glucose-6-phosphate. Has low glucose 1,6-bisphosphate synthase activity (By similarity).
Catalyzes the conversion of the nucleoside breakdown products ribose-1-phosphate and deoxyribose-1-phosphate to the corresponding 5-phosphopentoses. May also catalyze the interconversion of glucose-1-phosphate and glucose-6-phosphate. Has low glucose 1,6-bisphosphate synthase activity (By similarity).
Catalytic Activity
alpha-D-glucose 1-phosphate = alpha-D-glucose 6-phosphate
alpha-D-ribose 1-phosphate = D-ribose 5-phosphate
2-deoxy-alpha-D-ribose 1-phosphate = 2-deoxy-D-ribose 5-phosphate
alpha-D-ribose 1-phosphate = D-ribose 5-phosphate
2-deoxy-alpha-D-ribose 1-phosphate = 2-deoxy-D-ribose 5-phosphate
Cofactor
Mg(2+)
Biophysicochemical Properties
45.7 uM for alpha-D-ribose 1-phosphate
4.1 uM for 2-deoxy-alpha-D-ribose 1-phosphate
114 uM for alpha-D-glucose 1-phosphate
4.1 uM for 2-deoxy-alpha-D-ribose 1-phosphate
114 uM for alpha-D-glucose 1-phosphate
Similarity
Belongs to the phosphohexose mutase family.
Keywords
Acetylation
Alternative splicing
Carbohydrate metabolism
Complete proteome
Cytoplasm
Direct protein sequencing
Glucose metabolism
Isomerase
Magnesium
Metal-binding
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Phosphoglucomutase-2
splice variant In isoform 2.
sequence variant In dbSNP:rs17856324.
splice variant In isoform 2.
sequence variant In dbSNP:rs17856324.
Uniprot
A0A0L7LMT4
A0A2A4JE52
A0A2W1BQG6
A0A2H1VMC8
A0A3S2TPU5
A0A212ETF5
+ More
A0A194QN07 A0A194PJN5 H9JLP3 A0A3S3QJ92 A0A3S3PHL7 A0A2J7R8Q5 A0A2J7R8Q8 A0A1B6E2I8 A0A1Y1N3L1 A0A161ML37 A0A151PA90 A0A131XBP6 A0A210Q4X0 E2AE69 I3MB62 M3Y3H5 A0A2Y9JKR7 A0A023FLK0 B0XBU8 G9KG72 A0A293MVQ6 F7ENI5 F7B0S5 A0A3Q0HD21 A0A2R5LLS1 K7G6P2 K7G6P7 G3VSW5 E2BFH3 D2HTA4 A0A023F3Y4 A0A1S3EJ06 G1LXN4 A0A224Z1A7 L7MCS2 L7MEP4 A0A131YI42 A0A1B0D2T0 A0A0L8HIV4 K9KB11 A0A1Q3FKA7 A0A1Q3FKU7 A0A1V9YYT4 A0A0V0G6E7 L8Y7V8 F6SIC9 F6YK46 M7C5T9 B4DN40 A0A2U3XIL3 A0A1V9X8D7 A0A1L8E1J3 A0A182H7R3 K7ECI9 A0A182GXV6 A0A3Q2I8C1 Q95RR7 A0A2H6N7Z9 S7MK69 A0A3Q7XNU1 A0A0P4WDD6 A0A3Q2GWI8 A0A1Q3FJA4 A0A384CYM7 H9GCD8 G1PBJ6 A0A3Q7Q8Q7 A0A1Q3FKE8 A0A023GLE0 G3MPT2 A0A1B6K757 A0A2U3ZBK0 A0A0J7L3G8 A0A2Y9I4U3 A0A3Q7TDJ7 A0A2K5EMK0 F7FLB2 E2QV12 A0A2K6GR36 A0A1S3QMI8 A0A091UKI1 A0A2K6TGH8 A0A182T912 Q17L34 A0A0C9RME0 M3W2Q7 A0A091DQW1 G1S585 A0A2R9C685 A0A182RMC9 Q96G03 W5PJD0 H2PD34 Q5RFI8
A0A194QN07 A0A194PJN5 H9JLP3 A0A3S3QJ92 A0A3S3PHL7 A0A2J7R8Q5 A0A2J7R8Q8 A0A1B6E2I8 A0A1Y1N3L1 A0A161ML37 A0A151PA90 A0A131XBP6 A0A210Q4X0 E2AE69 I3MB62 M3Y3H5 A0A2Y9JKR7 A0A023FLK0 B0XBU8 G9KG72 A0A293MVQ6 F7ENI5 F7B0S5 A0A3Q0HD21 A0A2R5LLS1 K7G6P2 K7G6P7 G3VSW5 E2BFH3 D2HTA4 A0A023F3Y4 A0A1S3EJ06 G1LXN4 A0A224Z1A7 L7MCS2 L7MEP4 A0A131YI42 A0A1B0D2T0 A0A0L8HIV4 K9KB11 A0A1Q3FKA7 A0A1Q3FKU7 A0A1V9YYT4 A0A0V0G6E7 L8Y7V8 F6SIC9 F6YK46 M7C5T9 B4DN40 A0A2U3XIL3 A0A1V9X8D7 A0A1L8E1J3 A0A182H7R3 K7ECI9 A0A182GXV6 A0A3Q2I8C1 Q95RR7 A0A2H6N7Z9 S7MK69 A0A3Q7XNU1 A0A0P4WDD6 A0A3Q2GWI8 A0A1Q3FJA4 A0A384CYM7 H9GCD8 G1PBJ6 A0A3Q7Q8Q7 A0A1Q3FKE8 A0A023GLE0 G3MPT2 A0A1B6K757 A0A2U3ZBK0 A0A0J7L3G8 A0A2Y9I4U3 A0A3Q7TDJ7 A0A2K5EMK0 F7FLB2 E2QV12 A0A2K6GR36 A0A1S3QMI8 A0A091UKI1 A0A2K6TGH8 A0A182T912 Q17L34 A0A0C9RME0 M3W2Q7 A0A091DQW1 G1S585 A0A2R9C685 A0A182RMC9 Q96G03 W5PJD0 H2PD34 Q5RFI8
EC Number
5.4.2.2
Pubmed
26227816
28756777
22118469
26354079
19121390
28004739
+ More
22293439 28049606 28812685 20798317 23236062 17495919 17381049 21709235 20010809 25474469 28797301 25576852 26830274 25527045 23385571 18464734 19892987 23624526 28327890 26483478 24813606 21881562 21993624 22216098 15489334 15057822 22673903 16341006 17510324 17975172 22722832 11230166 14702039 15815621 12665801 17804405 18669648 19413330 20068231 21269460 22223895 22814378 23186163 24275569 20809919
22293439 28049606 28812685 20798317 23236062 17495919 17381049 21709235 20010809 25474469 28797301 25576852 26830274 25527045 23385571 18464734 19892987 23624526 28327890 26483478 24813606 21881562 21993624 22216098 15489334 15057822 22673903 16341006 17510324 17975172 22722832 11230166 14702039 15815621 12665801 17804405 18669648 19413330 20068231 21269460 22223895 22814378 23186163 24275569 20809919
EMBL
JTDY01000537
KOB76752.1
NWSH01001809
PCG70056.1
KZ150015
PZC74980.1
+ More
ODYU01003339 SOQ41967.1 RSAL01000028 RVE51871.1 AGBW02012613 OWR44724.1 KQ461194 KPJ06897.1 KQ459603 KPI92939.1 BABH01005300 NCKU01002418 NCKU01000946 NCKU01000944 RWS09628.1 RWS13567.1 RWS13577.1 NCKU01002341 RWS09801.1 NEVH01006721 PNF37212.1 PNF37214.1 GEDC01011087 GEDC01005156 JAS26211.1 JAS32142.1 GEZM01013572 GEZM01013571 GEZM01013570 GEZM01013569 JAV92473.1 GEMB01002571 JAS00618.1 AKHW03000533 KYO45884.1 GEFH01004192 JAP64389.1 NEDP02004995 OWF43792.1 GL438827 EFN68302.1 AGTP01015495 AEYP01059785 AEYP01059786 AEYP01059787 AEYP01059788 AEYP01059789 GBBK01002468 JAC22014.1 DS232654 EDS44477.1 JP015299 AES03897.1 GFWV01020205 MAA44933.1 GGLE01006313 MBY10439.1 AGCU01130327 AGCU01130328 AGCU01130329 AGCU01130330 AEFK01223666 GL448037 EFN85495.1 GL193334 EFB18287.1 GBBI01003048 JAC15664.1 ACTA01059617 ACTA01067617 ACTA01075617 GFPF01010583 MAA21729.1 GACK01003244 JAA61790.1 GACK01003306 JAA61728.1 GEDV01009638 JAP78919.1 AJVK01002791 KQ418031 KOF89193.1 JL619537 AEP99170.1 GFDL01007129 JAV27916.1 GFDL01006890 JAV28155.1 JNBR01000567 OQR90918.1 GECL01002547 JAP03577.1 KB367707 ELV10446.1 KB527356 EMP35877.1 AK297752 BAG60102.1 MNPL01019665 OQR69827.1 GFDF01001484 JAV12600.1 JXUM01004201 JXUM01004202 KQ560170 KXJ84032.1 JXUM01096349 KQ564258 KXJ72517.1 AY061184 AAL28732.2 IACI01044885 LAA24465.1 KE161555 EPQ04481.1 GDRN01067212 JAI64399.1 GFDL01007334 JAV27711.1 AAWZ02026073 AAPE02017312 GFDL01006988 JAV28057.1 GBBM01000686 JAC34732.1 JO843883 AEO35500.1 GECU01000427 JAT07280.1 LBMM01000816 KMQ97422.1 AABR07015025 AABR07015026 BC160893 AAI60893.1 AAEX03002546 KK455876 KFQ76273.1 CH477219 EAT47392.1 GBYB01014567 JAG84334.1 AANG04001605 KN122111 KFO32853.1 ADFV01182385 ADFV01182386 ADFV01182387 ADFV01182388 AJFE02114342 AJFE02114343 AL136705 AF109360 AK001845 AK303374 CR457274 AK223237 AC021106 AC108022 BC010087 AMGL01098034 ABGA01329922 ABGA01329923 ABGA01329924 ABGA01329925 NDHI03003508 PNJ30749.1 CR857168
ODYU01003339 SOQ41967.1 RSAL01000028 RVE51871.1 AGBW02012613 OWR44724.1 KQ461194 KPJ06897.1 KQ459603 KPI92939.1 BABH01005300 NCKU01002418 NCKU01000946 NCKU01000944 RWS09628.1 RWS13567.1 RWS13577.1 NCKU01002341 RWS09801.1 NEVH01006721 PNF37212.1 PNF37214.1 GEDC01011087 GEDC01005156 JAS26211.1 JAS32142.1 GEZM01013572 GEZM01013571 GEZM01013570 GEZM01013569 JAV92473.1 GEMB01002571 JAS00618.1 AKHW03000533 KYO45884.1 GEFH01004192 JAP64389.1 NEDP02004995 OWF43792.1 GL438827 EFN68302.1 AGTP01015495 AEYP01059785 AEYP01059786 AEYP01059787 AEYP01059788 AEYP01059789 GBBK01002468 JAC22014.1 DS232654 EDS44477.1 JP015299 AES03897.1 GFWV01020205 MAA44933.1 GGLE01006313 MBY10439.1 AGCU01130327 AGCU01130328 AGCU01130329 AGCU01130330 AEFK01223666 GL448037 EFN85495.1 GL193334 EFB18287.1 GBBI01003048 JAC15664.1 ACTA01059617 ACTA01067617 ACTA01075617 GFPF01010583 MAA21729.1 GACK01003244 JAA61790.1 GACK01003306 JAA61728.1 GEDV01009638 JAP78919.1 AJVK01002791 KQ418031 KOF89193.1 JL619537 AEP99170.1 GFDL01007129 JAV27916.1 GFDL01006890 JAV28155.1 JNBR01000567 OQR90918.1 GECL01002547 JAP03577.1 KB367707 ELV10446.1 KB527356 EMP35877.1 AK297752 BAG60102.1 MNPL01019665 OQR69827.1 GFDF01001484 JAV12600.1 JXUM01004201 JXUM01004202 KQ560170 KXJ84032.1 JXUM01096349 KQ564258 KXJ72517.1 AY061184 AAL28732.2 IACI01044885 LAA24465.1 KE161555 EPQ04481.1 GDRN01067212 JAI64399.1 GFDL01007334 JAV27711.1 AAWZ02026073 AAPE02017312 GFDL01006988 JAV28057.1 GBBM01000686 JAC34732.1 JO843883 AEO35500.1 GECU01000427 JAT07280.1 LBMM01000816 KMQ97422.1 AABR07015025 AABR07015026 BC160893 AAI60893.1 AAEX03002546 KK455876 KFQ76273.1 CH477219 EAT47392.1 GBYB01014567 JAG84334.1 AANG04001605 KN122111 KFO32853.1 ADFV01182385 ADFV01182386 ADFV01182387 ADFV01182388 AJFE02114342 AJFE02114343 AL136705 AF109360 AK001845 AK303374 CR457274 AK223237 AC021106 AC108022 BC010087 AMGL01098034 ABGA01329922 ABGA01329923 ABGA01329924 ABGA01329925 NDHI03003508 PNJ30749.1 CR857168
Proteomes
UP000037510
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000005204 UP000285301 UP000235965 UP000050525 UP000242188 UP000000311 UP000005215 UP000000715 UP000248482 UP000002320 UP000002280 UP000189705 UP000007267 UP000007648 UP000008237 UP000081671 UP000008912 UP000092462 UP000053454 UP000243579 UP000011518 UP000002279 UP000002281 UP000031443 UP000245341 UP000192247 UP000069940 UP000249989 UP000286642 UP000261680 UP000291021 UP000001646 UP000001074 UP000286641 UP000245340 UP000036403 UP000248481 UP000286640 UP000233020 UP000002494 UP000002254 UP000233160 UP000087266 UP000233220 UP000075901 UP000008820 UP000011712 UP000028990 UP000001073 UP000240080 UP000075900 UP000005640 UP000002356 UP000001595
UP000005204 UP000285301 UP000235965 UP000050525 UP000242188 UP000000311 UP000005215 UP000000715 UP000248482 UP000002320 UP000002280 UP000189705 UP000007267 UP000007648 UP000008237 UP000081671 UP000008912 UP000092462 UP000053454 UP000243579 UP000011518 UP000002279 UP000002281 UP000031443 UP000245341 UP000192247 UP000069940 UP000249989 UP000286642 UP000261680 UP000291021 UP000001646 UP000001074 UP000286641 UP000245340 UP000036403 UP000248481 UP000286640 UP000233020 UP000002494 UP000002254 UP000233160 UP000087266 UP000233220 UP000075901 UP000008820 UP000011712 UP000028990 UP000001073 UP000240080 UP000075900 UP000005640 UP000002356 UP000001595
Interpro
ProteinModelPortal
A0A0L7LMT4
A0A2A4JE52
A0A2W1BQG6
A0A2H1VMC8
A0A3S2TPU5
A0A212ETF5
+ More
A0A194QN07 A0A194PJN5 H9JLP3 A0A3S3QJ92 A0A3S3PHL7 A0A2J7R8Q5 A0A2J7R8Q8 A0A1B6E2I8 A0A1Y1N3L1 A0A161ML37 A0A151PA90 A0A131XBP6 A0A210Q4X0 E2AE69 I3MB62 M3Y3H5 A0A2Y9JKR7 A0A023FLK0 B0XBU8 G9KG72 A0A293MVQ6 F7ENI5 F7B0S5 A0A3Q0HD21 A0A2R5LLS1 K7G6P2 K7G6P7 G3VSW5 E2BFH3 D2HTA4 A0A023F3Y4 A0A1S3EJ06 G1LXN4 A0A224Z1A7 L7MCS2 L7MEP4 A0A131YI42 A0A1B0D2T0 A0A0L8HIV4 K9KB11 A0A1Q3FKA7 A0A1Q3FKU7 A0A1V9YYT4 A0A0V0G6E7 L8Y7V8 F6SIC9 F6YK46 M7C5T9 B4DN40 A0A2U3XIL3 A0A1V9X8D7 A0A1L8E1J3 A0A182H7R3 K7ECI9 A0A182GXV6 A0A3Q2I8C1 Q95RR7 A0A2H6N7Z9 S7MK69 A0A3Q7XNU1 A0A0P4WDD6 A0A3Q2GWI8 A0A1Q3FJA4 A0A384CYM7 H9GCD8 G1PBJ6 A0A3Q7Q8Q7 A0A1Q3FKE8 A0A023GLE0 G3MPT2 A0A1B6K757 A0A2U3ZBK0 A0A0J7L3G8 A0A2Y9I4U3 A0A3Q7TDJ7 A0A2K5EMK0 F7FLB2 E2QV12 A0A2K6GR36 A0A1S3QMI8 A0A091UKI1 A0A2K6TGH8 A0A182T912 Q17L34 A0A0C9RME0 M3W2Q7 A0A091DQW1 G1S585 A0A2R9C685 A0A182RMC9 Q96G03 W5PJD0 H2PD34 Q5RFI8
A0A194QN07 A0A194PJN5 H9JLP3 A0A3S3QJ92 A0A3S3PHL7 A0A2J7R8Q5 A0A2J7R8Q8 A0A1B6E2I8 A0A1Y1N3L1 A0A161ML37 A0A151PA90 A0A131XBP6 A0A210Q4X0 E2AE69 I3MB62 M3Y3H5 A0A2Y9JKR7 A0A023FLK0 B0XBU8 G9KG72 A0A293MVQ6 F7ENI5 F7B0S5 A0A3Q0HD21 A0A2R5LLS1 K7G6P2 K7G6P7 G3VSW5 E2BFH3 D2HTA4 A0A023F3Y4 A0A1S3EJ06 G1LXN4 A0A224Z1A7 L7MCS2 L7MEP4 A0A131YI42 A0A1B0D2T0 A0A0L8HIV4 K9KB11 A0A1Q3FKA7 A0A1Q3FKU7 A0A1V9YYT4 A0A0V0G6E7 L8Y7V8 F6SIC9 F6YK46 M7C5T9 B4DN40 A0A2U3XIL3 A0A1V9X8D7 A0A1L8E1J3 A0A182H7R3 K7ECI9 A0A182GXV6 A0A3Q2I8C1 Q95RR7 A0A2H6N7Z9 S7MK69 A0A3Q7XNU1 A0A0P4WDD6 A0A3Q2GWI8 A0A1Q3FJA4 A0A384CYM7 H9GCD8 G1PBJ6 A0A3Q7Q8Q7 A0A1Q3FKE8 A0A023GLE0 G3MPT2 A0A1B6K757 A0A2U3ZBK0 A0A0J7L3G8 A0A2Y9I4U3 A0A3Q7TDJ7 A0A2K5EMK0 F7FLB2 E2QV12 A0A2K6GR36 A0A1S3QMI8 A0A091UKI1 A0A2K6TGH8 A0A182T912 Q17L34 A0A0C9RME0 M3W2Q7 A0A091DQW1 G1S585 A0A2R9C685 A0A182RMC9 Q96G03 W5PJD0 H2PD34 Q5RFI8
Ontologies
GO
GO:0016868
GO:0005975
GO:0000287
GO:0046872
GO:0008973
GO:0006006
GO:0004614
GO:0016021
GO:0005576
GO:0070062
GO:0034774
GO:0046386
GO:0006098
GO:0019388
GO:0005829
GO:0005980
GO:1904813
GO:0005978
GO:0043312
GO:0005737
GO:0008081
GO:0005634
GO:0008270
GO:0005515
GO:0004319
GO:0004320
GO:0016295
GO:0016296
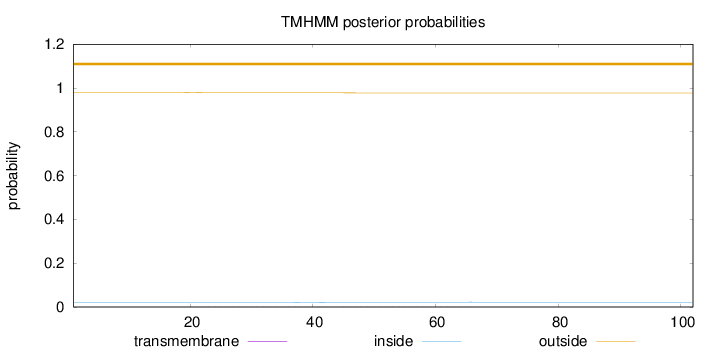
Topology
Subcellular location
Cytoplasm
Length:
102
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0285
Exp number, first 60 AAs:
0.02477
Total prob of N-in:
0.02083
outside
1 - 102
Population Genetic Test Statistics
Pi
149.251094
Theta
74.416648
Tajima's D
0.312721
CLR
0.037307
CSRT
0.468376581170941
Interpretation
Uncertain
