Gene
KWMTBOMO07061 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010445
Annotation
PREDICTED:_phosphoglucomutase-2_[Amyelois_transitella]
Full name
Glucose 1,6-bisphosphate synthase
Alternative Name
Phosphoglucomutase-2-like 1
Location in the cell
Extracellular Reliability : 1.409 Nuclear Reliability : 1.888
Sequence
CDS
ATGTCGCTAGGCTCCCAGCTGCAGCGGCTCTACGCGGAGTACGGGTACCACGTGTCGCACAACTCCTACTACATCTGCCACGAGCCCTCCACCATACAGAAGATCTTCAGGAGGATCAGGAATTACGAGGGACCCGGTCAATACCCGCGGCGGGTGGGCTCGTGCGGCGTCACGCGCGTCACGGACTTCACGGCCGGCGTCACGGCGCCCCCCGCGCCCCGCACCGATACCCACCTACGCGTGCTAGGCACCGGGCTGGACCAGGACTACACGAGCGGCGAGATGGTGATGTTCCGCTGCGACACGGGGCTGAGCGCGTGCGTGCGCACCAGCGGGACCGAGCCCAAGCTCAAGTACTACACCGAGCTGGTCTGCCGGGCCGACACCGTCAGTGAACAAGAGGAAATGAAATCGAAACTAAAGGAGATCGTTCAAAAGTTCATCAATGAGCTCCTACAGCCCGAAGAGAACGGTCTGATAGGATAA
Protein
MSLGSQLQRLYAEYGYHVSHNSYYICHEPSTIQKIFRRIRNYEGPGQYPRRVGSCGVTRVTDFTAGVTAPPAPRTDTHLRVLGTGLDQDYTSGEMVMFRCDTGLSACVRTSGTEPKLKYYTELVCRADTVSEQEEMKSKLKEIVQKFINELLQPEENGLIG
Summary
Description
Glucose 1,6-bisphosphate synthase using 1,3-bisphosphoglycerate as a phosphate donor and a series of 1-phosphate sugars as acceptors, including glucose 1-phosphate, mannose 1-phosphate, ribose 1-phosphate and deoxyribose 1-phosphate. 5 or 6-phosphosugars are bad substrates, with the exception of glucose 6-phosphate. Also synthesizes ribose 1,5-bisphosphate. Has only low phosphopentomutase and phosphoglucomutase activities (By similarity).
Catalytic Activity
3-phospho-D-glyceroyl phosphate + alpha-D-glucose 1-phosphate = 3-phospho-D-glycerate + alpha-D-glucose 1,6-bisphosphate + H(+)
Similarity
Belongs to the phosphohexose mutase family.
Keywords
Carbohydrate metabolism
Complete proteome
Glucose metabolism
Isomerase
Magnesium
Metal-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain Glucose 1,6-bisphosphate synthase
Uniprot
A0A2H1VMC8
A0A194QN07
A0A2W1BQG6
A0A2A4JE52
A0A3S2TPU5
A0A212ETF5
+ More
A0A1Y1N3L1 A0A1S3JHS0 V9KQM6 A0A3B4ANJ1 A0A3Q2W2Q3 A0A3B4FUX8 A0A2U9B6W8 A0A0F8AFZ1 A0A087XY92 A0A3Q2ZR41 A0A3Q1ET40 A0A3Q0JH07 A0A3R7MHH8 A0A3B3UL15 A0A1A8Q3U5 A0A3P4S5T8 H2ZU38 W5N5G0 A0A3P8PDD7 A0A1A8MME7 A0A3P9AWN6 I3KKS9 A0A131YI42 A0A2K5UVA4 A0A3P8TNI5 G7PN84 I2CTQ0 H9ET98 A0A3N0XFB5 A0A2K5UV16 A0A1W0WP91 A0A224Z1A7 A0A2R8MNV7 A0A1B6K757 K9KEX3 A0A026WDR6 A0A3L8E3D6 L7MCS2 A0A3Q1CE91 A0A3Q3N7B3 L7MEP4 A0A2R8N2I2 A0A3P9MY55 A0A1A8HI91 A0A3B1JXH5 A0A3P9HUR8 A0A1D2NCZ4 A0A2P4SYM6 F6UD41 A0A3Q3W7A0 A0A3B4TRD3 A0A3Q3MYU5 A0A154PI01 G3UV94 A7RVP4 A0A1A8A2E5 A0A1A8U4C2 A0A3B4YUC4 A0A2K5LDS2 A0A2K6BMT4 A0A2K6MV95 A0A183PHU3 A0A1B6E2I8 A0A2I3N9Q7 T1JNB8 A0A2K6AKN3 A0A2R9A428 A0A2I3SH55 A0A3Q1JG52 A0A340WKM0 A0A2K6SP83 A0A131XBP6 A0A2K6MVA0 A0A2K6Q0U5 A0A0S7KN38 A0A0D9QVI1 A0A2K5LDP4 A0A2K6MV92 A0A3Q2DGL1 A0A2K6BMI4 A0A2I2YPZ7 A0A2K6AKP4 A0A2R9A9G5 A0A096MUB3 A0A2J8V1E3 H2Q4E1 Q5R979 A0A2K6Q0V0 A0A1A8DEP7 G3QXX0 A0A3Q0SYG3 A0A2I3GIA9
A0A1Y1N3L1 A0A1S3JHS0 V9KQM6 A0A3B4ANJ1 A0A3Q2W2Q3 A0A3B4FUX8 A0A2U9B6W8 A0A0F8AFZ1 A0A087XY92 A0A3Q2ZR41 A0A3Q1ET40 A0A3Q0JH07 A0A3R7MHH8 A0A3B3UL15 A0A1A8Q3U5 A0A3P4S5T8 H2ZU38 W5N5G0 A0A3P8PDD7 A0A1A8MME7 A0A3P9AWN6 I3KKS9 A0A131YI42 A0A2K5UVA4 A0A3P8TNI5 G7PN84 I2CTQ0 H9ET98 A0A3N0XFB5 A0A2K5UV16 A0A1W0WP91 A0A224Z1A7 A0A2R8MNV7 A0A1B6K757 K9KEX3 A0A026WDR6 A0A3L8E3D6 L7MCS2 A0A3Q1CE91 A0A3Q3N7B3 L7MEP4 A0A2R8N2I2 A0A3P9MY55 A0A1A8HI91 A0A3B1JXH5 A0A3P9HUR8 A0A1D2NCZ4 A0A2P4SYM6 F6UD41 A0A3Q3W7A0 A0A3B4TRD3 A0A3Q3MYU5 A0A154PI01 G3UV94 A7RVP4 A0A1A8A2E5 A0A1A8U4C2 A0A3B4YUC4 A0A2K5LDS2 A0A2K6BMT4 A0A2K6MV95 A0A183PHU3 A0A1B6E2I8 A0A2I3N9Q7 T1JNB8 A0A2K6AKN3 A0A2R9A428 A0A2I3SH55 A0A3Q1JG52 A0A340WKM0 A0A2K6SP83 A0A131XBP6 A0A2K6MVA0 A0A2K6Q0U5 A0A0S7KN38 A0A0D9QVI1 A0A2K5LDP4 A0A2K6MV92 A0A3Q2DGL1 A0A2K6BMI4 A0A2I2YPZ7 A0A2K6AKP4 A0A2R9A9G5 A0A096MUB3 A0A2J8V1E3 H2Q4E1 Q5R979 A0A2K6Q0V0 A0A1A8DEP7 G3QXX0 A0A3Q0SYG3 A0A2I3GIA9
EC Number
2.7.1.106
Pubmed
EMBL
ODYU01003339
SOQ41967.1
KQ461194
KPJ06897.1
KZ150015
PZC74980.1
+ More
NWSH01001809 PCG70056.1 RSAL01000028 RVE51871.1 AGBW02012613 OWR44724.1 GEZM01013572 GEZM01013571 GEZM01013570 GEZM01013569 JAV92473.1 JW868127 AFP00645.1 CP026245 AWO99511.1 KQ042224 KKF17480.1 AYCK01015536 QCYY01001620 ROT76706.1 HAEH01009736 SBR87922.1 CYRY02046764 VCX42560.1 AFYH01231781 AFYH01231782 AFYH01231783 AFYH01231784 AFYH01231785 AHAT01001929 AHAT01001930 AHAT01001931 HAEF01016582 SBR57741.1 AERX01047717 GEDV01009638 JAP78919.1 AQIA01021407 AQIA01021408 CM001289 EHH56579.1 JV565000 CM001266 AFJ70335.1 EHH23243.1 JU321851 JU473577 AFE65607.1 AFH30381.1 RJVU01075617 ROI16020.1 MTYJ01000067 OQV17026.1 GFPF01010583 MAA21729.1 GECU01000427 JAT07280.1 JL624856 AEP99871.1 KK107260 EZA54197.1 QOIP01000001 RLU27260.1 GACK01003244 JAA61790.1 GACK01003306 JAA61728.1 HAEC01015724 SBQ83945.1 LJIJ01000086 ODN03111.1 PPHD01016190 POI29208.1 GAMS01003426 GAMS01003425 GAMS01003424 GAMS01003423 GAMR01004752 GAMR01004751 GAMR01004750 GAMR01004749 GAMR01004748 GAMP01002974 JAB19710.1 JAB29180.1 JAB49781.1 KQ434899 KZC10948.1 DS469544 EDO44486.1 HADY01010338 SBP48823.1 HAEJ01001979 SBS42436.1 UZAL01034062 VDP64622.1 GEDC01011087 GEDC01005156 JAS26211.1 JAS32142.1 AHZZ02007147 AHZZ02007148 AHZZ02007149 JH431376 AJFE02047950 AJFE02047951 AJFE02047952 AACZ04016281 GEFH01004192 JAP64389.1 GBYX01203502 GBYX01203495 JAO78720.1 AQIB01134126 AQIB01134127 CABD030080217 CABD030080218 CABD030080219 CABD030080220 CABD030080221 CABD030080222 NDHI03003437 PNJ51341.1 GABC01008945 GABF01003583 GABE01004996 NBAG03000261 JAA02393.1 JAA18562.1 JAA39743.1 PNI56681.1 CR859079 CR859513 HAEA01003408 SBQ31888.1 ADFV01111549 ADFV01111550 ADFV01111551 ADFV01111552 ADFV01111553
NWSH01001809 PCG70056.1 RSAL01000028 RVE51871.1 AGBW02012613 OWR44724.1 GEZM01013572 GEZM01013571 GEZM01013570 GEZM01013569 JAV92473.1 JW868127 AFP00645.1 CP026245 AWO99511.1 KQ042224 KKF17480.1 AYCK01015536 QCYY01001620 ROT76706.1 HAEH01009736 SBR87922.1 CYRY02046764 VCX42560.1 AFYH01231781 AFYH01231782 AFYH01231783 AFYH01231784 AFYH01231785 AHAT01001929 AHAT01001930 AHAT01001931 HAEF01016582 SBR57741.1 AERX01047717 GEDV01009638 JAP78919.1 AQIA01021407 AQIA01021408 CM001289 EHH56579.1 JV565000 CM001266 AFJ70335.1 EHH23243.1 JU321851 JU473577 AFE65607.1 AFH30381.1 RJVU01075617 ROI16020.1 MTYJ01000067 OQV17026.1 GFPF01010583 MAA21729.1 GECU01000427 JAT07280.1 JL624856 AEP99871.1 KK107260 EZA54197.1 QOIP01000001 RLU27260.1 GACK01003244 JAA61790.1 GACK01003306 JAA61728.1 HAEC01015724 SBQ83945.1 LJIJ01000086 ODN03111.1 PPHD01016190 POI29208.1 GAMS01003426 GAMS01003425 GAMS01003424 GAMS01003423 GAMR01004752 GAMR01004751 GAMR01004750 GAMR01004749 GAMR01004748 GAMP01002974 JAB19710.1 JAB29180.1 JAB49781.1 KQ434899 KZC10948.1 DS469544 EDO44486.1 HADY01010338 SBP48823.1 HAEJ01001979 SBS42436.1 UZAL01034062 VDP64622.1 GEDC01011087 GEDC01005156 JAS26211.1 JAS32142.1 AHZZ02007147 AHZZ02007148 AHZZ02007149 JH431376 AJFE02047950 AJFE02047951 AJFE02047952 AACZ04016281 GEFH01004192 JAP64389.1 GBYX01203502 GBYX01203495 JAO78720.1 AQIB01134126 AQIB01134127 CABD030080217 CABD030080218 CABD030080219 CABD030080220 CABD030080221 CABD030080222 NDHI03003437 PNJ51341.1 GABC01008945 GABF01003583 GABE01004996 NBAG03000261 JAA02393.1 JAA18562.1 JAA39743.1 PNI56681.1 CR859079 CR859513 HAEA01003408 SBQ31888.1 ADFV01111549 ADFV01111550 ADFV01111551 ADFV01111552 ADFV01111553
Proteomes
UP000053240
UP000218220
UP000283053
UP000007151
UP000085678
UP000261520
+ More
UP000264840 UP000261460 UP000246464 UP000028760 UP000264800 UP000257200 UP000079169 UP000283509 UP000261500 UP000008672 UP000018468 UP000265100 UP000265160 UP000005207 UP000233100 UP000265080 UP000009130 UP000008225 UP000053097 UP000279307 UP000257160 UP000261640 UP000242638 UP000018467 UP000265200 UP000094527 UP000261620 UP000261420 UP000076502 UP000001645 UP000001593 UP000261360 UP000233060 UP000233120 UP000233180 UP000050791 UP000028761 UP000233140 UP000240080 UP000002277 UP000265040 UP000265300 UP000233220 UP000233200 UP000029965 UP000265020 UP000001519 UP000001595 UP000261340 UP000001073
UP000264840 UP000261460 UP000246464 UP000028760 UP000264800 UP000257200 UP000079169 UP000283509 UP000261500 UP000008672 UP000018468 UP000265100 UP000265160 UP000005207 UP000233100 UP000265080 UP000009130 UP000008225 UP000053097 UP000279307 UP000257160 UP000261640 UP000242638 UP000018467 UP000265200 UP000094527 UP000261620 UP000261420 UP000076502 UP000001645 UP000001593 UP000261360 UP000233060 UP000233120 UP000233180 UP000050791 UP000028761 UP000233140 UP000240080 UP000002277 UP000265040 UP000265300 UP000233220 UP000233200 UP000029965 UP000265020 UP000001519 UP000001595 UP000261340 UP000001073
Pfam
Interpro
IPR005844
A-D-PHexomutase_a/b/a-I
+ More
IPR005845 A-D-PHexomutase_a/b/a-II
IPR036900 A-D-PHexomutase_C_sf
IPR005846 A-D-PHexomutase_a/b/a-III
IPR016066 A-D-PHexomutase_CS
IPR016055 A-D-PHexomutase_a/b/a-I/II/III
IPR005841 Alpha-D-phosphohexomutase_SF
IPR005843 A-D-PHexomutase_C
IPR038386 Beta-thymosin_sf
IPR001152 Beta-thymosin
IPR005845 A-D-PHexomutase_a/b/a-II
IPR036900 A-D-PHexomutase_C_sf
IPR005846 A-D-PHexomutase_a/b/a-III
IPR016066 A-D-PHexomutase_CS
IPR016055 A-D-PHexomutase_a/b/a-I/II/III
IPR005841 Alpha-D-phosphohexomutase_SF
IPR005843 A-D-PHexomutase_C
IPR038386 Beta-thymosin_sf
IPR001152 Beta-thymosin
Gene 3D
ProteinModelPortal
A0A2H1VMC8
A0A194QN07
A0A2W1BQG6
A0A2A4JE52
A0A3S2TPU5
A0A212ETF5
+ More
A0A1Y1N3L1 A0A1S3JHS0 V9KQM6 A0A3B4ANJ1 A0A3Q2W2Q3 A0A3B4FUX8 A0A2U9B6W8 A0A0F8AFZ1 A0A087XY92 A0A3Q2ZR41 A0A3Q1ET40 A0A3Q0JH07 A0A3R7MHH8 A0A3B3UL15 A0A1A8Q3U5 A0A3P4S5T8 H2ZU38 W5N5G0 A0A3P8PDD7 A0A1A8MME7 A0A3P9AWN6 I3KKS9 A0A131YI42 A0A2K5UVA4 A0A3P8TNI5 G7PN84 I2CTQ0 H9ET98 A0A3N0XFB5 A0A2K5UV16 A0A1W0WP91 A0A224Z1A7 A0A2R8MNV7 A0A1B6K757 K9KEX3 A0A026WDR6 A0A3L8E3D6 L7MCS2 A0A3Q1CE91 A0A3Q3N7B3 L7MEP4 A0A2R8N2I2 A0A3P9MY55 A0A1A8HI91 A0A3B1JXH5 A0A3P9HUR8 A0A1D2NCZ4 A0A2P4SYM6 F6UD41 A0A3Q3W7A0 A0A3B4TRD3 A0A3Q3MYU5 A0A154PI01 G3UV94 A7RVP4 A0A1A8A2E5 A0A1A8U4C2 A0A3B4YUC4 A0A2K5LDS2 A0A2K6BMT4 A0A2K6MV95 A0A183PHU3 A0A1B6E2I8 A0A2I3N9Q7 T1JNB8 A0A2K6AKN3 A0A2R9A428 A0A2I3SH55 A0A3Q1JG52 A0A340WKM0 A0A2K6SP83 A0A131XBP6 A0A2K6MVA0 A0A2K6Q0U5 A0A0S7KN38 A0A0D9QVI1 A0A2K5LDP4 A0A2K6MV92 A0A3Q2DGL1 A0A2K6BMI4 A0A2I2YPZ7 A0A2K6AKP4 A0A2R9A9G5 A0A096MUB3 A0A2J8V1E3 H2Q4E1 Q5R979 A0A2K6Q0V0 A0A1A8DEP7 G3QXX0 A0A3Q0SYG3 A0A2I3GIA9
A0A1Y1N3L1 A0A1S3JHS0 V9KQM6 A0A3B4ANJ1 A0A3Q2W2Q3 A0A3B4FUX8 A0A2U9B6W8 A0A0F8AFZ1 A0A087XY92 A0A3Q2ZR41 A0A3Q1ET40 A0A3Q0JH07 A0A3R7MHH8 A0A3B3UL15 A0A1A8Q3U5 A0A3P4S5T8 H2ZU38 W5N5G0 A0A3P8PDD7 A0A1A8MME7 A0A3P9AWN6 I3KKS9 A0A131YI42 A0A2K5UVA4 A0A3P8TNI5 G7PN84 I2CTQ0 H9ET98 A0A3N0XFB5 A0A2K5UV16 A0A1W0WP91 A0A224Z1A7 A0A2R8MNV7 A0A1B6K757 K9KEX3 A0A026WDR6 A0A3L8E3D6 L7MCS2 A0A3Q1CE91 A0A3Q3N7B3 L7MEP4 A0A2R8N2I2 A0A3P9MY55 A0A1A8HI91 A0A3B1JXH5 A0A3P9HUR8 A0A1D2NCZ4 A0A2P4SYM6 F6UD41 A0A3Q3W7A0 A0A3B4TRD3 A0A3Q3MYU5 A0A154PI01 G3UV94 A7RVP4 A0A1A8A2E5 A0A1A8U4C2 A0A3B4YUC4 A0A2K5LDS2 A0A2K6BMT4 A0A2K6MV95 A0A183PHU3 A0A1B6E2I8 A0A2I3N9Q7 T1JNB8 A0A2K6AKN3 A0A2R9A428 A0A2I3SH55 A0A3Q1JG52 A0A340WKM0 A0A2K6SP83 A0A131XBP6 A0A2K6MVA0 A0A2K6Q0U5 A0A0S7KN38 A0A0D9QVI1 A0A2K5LDP4 A0A2K6MV92 A0A3Q2DGL1 A0A2K6BMI4 A0A2I2YPZ7 A0A2K6AKP4 A0A2R9A9G5 A0A096MUB3 A0A2J8V1E3 H2Q4E1 Q5R979 A0A2K6Q0V0 A0A1A8DEP7 G3QXX0 A0A3Q0SYG3 A0A2I3GIA9
Ontologies
GO
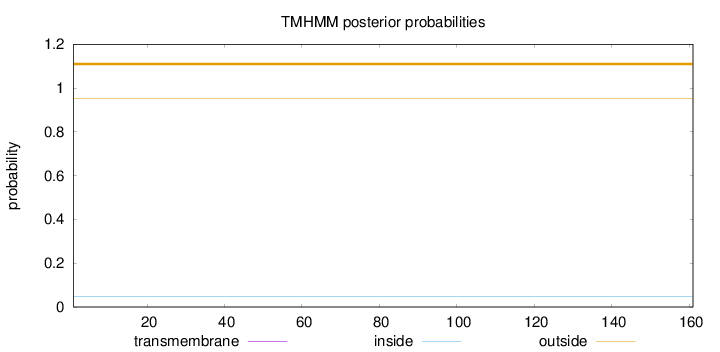
Topology
Length:
161
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00074
Exp number, first 60 AAs:
0.00074
Total prob of N-in:
0.04813
outside
1 - 161
Population Genetic Test Statistics
Pi
251.263027
Theta
192.049788
Tajima's D
0.018419
CLR
0.238597
CSRT
0.370531473426329
Interpretation
Uncertain
