Gene
KWMTBOMO07057 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010520
Annotation
PREDICTED:_ribonuclease_Oy_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.395 Extracellular Reliability : 1.495
Sequence
CDS
ATGTGGAGAAATTTGAGTTATGCTATATTTTTGCTGTCGTGTTTTACATATGCAGAACTAAGCGACACAAAAAATAATGAGTGGGATTTTCTAATATTCACGCAACACTGGCCGGCGACTATATGCAGAATATGGATGGAGAAACCTACGCACTCTTGTATTATGCCACACGAGCAGAATTCATGGACTATTCATGGTATATGGCCAAATAAAAATCACTCCATGGGACCATTTTTCTGTAATAAGACTTGGTCATTTGACCCTGATCAAGTTAAATCTATTGAAACAGACTTGGAAGAATCTTGGGGCAATATTGAGAAAGAACAATCATTGTACAGTTTATGGGCCCACGAATGGACAAAACACGGTACTTGCGCCGCATCAATCGAGCCCCTTAACTGCGAGCTGAAATACTTCACGAAAGGGTTAGAATGGCTGAAAAAGTATACCATGAAAAATATTTTGAACAATGCAGGTATCACCCCTTCAGACATGAAAGAGTACAAAGCTATTGATTTTCATAATGCTGTGAAAACTAAACTTGGATTTAAACCAATGGTTGAATGTCACAAAGTTGACGGTAAACAATTCTTGTTTGAAATACGCATATGCTTCTCAAAGTCACTTGAATTGATTGACTGTGAAGATACACATATGACTAATACGGATATTATTACGAATTGTAAGCCAACTGAAGGGATTATATATCCGGTAAATGAGAAACCTCCCAAAAGATATTTGATACAGCTTCATAGATTATTGTCATGGTTACGTTGGTTCACAGTGTAA
Protein
MWRNLSYAIFLLSCFTYAELSDTKNNEWDFLIFTQHWPATICRIWMEKPTHSCIMPHEQNSWTIHGIWPNKNHSMGPFFCNKTWSFDPDQVKSIETDLEESWGNIEKEQSLYSLWAHEWTKHGTCAASIEPLNCELKYFTKGLEWLKKYTMKNILNNAGITPSDMKEYKAIDFHNAVKTKLGFKPMVECHKVDGKQFLFEIRICFSKSLELIDCEDTHMTNTDIITNCKPTEGIIYPVNEKPPKRYLIQLHRLLSWLRWFTV
Summary
Similarity
Belongs to the RNase T2 family.
Uniprot
H9JLW8
A0A2H1VND0
A0A2W1BMY3
A0A2A4J987
A0A3S2LQR4
A0A194QNG1
+ More
A0A194PIR6 I4DQ25 A0A212ETF2 A0A0L7KA08 S4PX45 A0A1B6DBJ7 A0A088AN65 A0A1B0CEA7 W8B448 A0A2A4J9C6 A0A2P8Z3D4 A0A1B6MA73 A0A2W1BTC8 A0A1B6I0C2 A0A1B6JJ87 B4KUP5 A0A0A1XLT4 A0A1J1I4U7 A0A336MN48 E9GQD7 B4LHS3 B4PCD0 D3TLW3 A0A1B0DAP6 A0A1L8DPQ1 B4QKZ0 A0A0L0BPC5 A0A1A9ZMR2 A0A1L8DXL4 U5EP87 B4HIX6 Q95TA4 A0A1W4VNI4 A0A0P5FWM6 A0A0P6AGE2 A0A0P6JJU3 Q9VSC3 Q24485 Q5U199 A0A336M662 B3NF74 A0A0P5M3G1 A0A2J7PTZ5 A0A067RJE8 A0A0P5LAE3 A0A0P5GJK9 A0A034VRC3 A0A0N8CV52 A0A0M4EJW8 B4J107 A0A0N1ITF6 A0A0K8WAA3 A0A0P5C0W7 A0A0N8A6Q3 A0A1A9WCE6 A0A1A9UVT1 Q29EP9 A0A1B6FQF4 A0A1Q3FNT2 A0A0L7RDF1 F4W570 B4H2B9 B3M5P6 Q1HQZ4 A0A1W4X0F2 A0A3B0K2R9 A0A158NYY0 A0A023ENC4 A0A1B0AQ50 A0A2A3EE73 A0A1W6EWB1 A0A1A9XTW2 B4N5I7 A0A2M3Z635 A0A1S3IDK3 A0A2M3Z6X1 B0WUA5 T1PE88 A0A1I8M8I6 A0A2M4AIE7 A0A2M4AI59 T1GHR4 W5J922 A0A195D0W7 A0A0T6AVS1 A0A0P4VI06 A0A0A9Z961 A0A146KWQ2 A0A0V0G625 E9IUX5 T1IEB5 A0A224XGW2 A0A232EJ98 U4UBW7
A0A194PIR6 I4DQ25 A0A212ETF2 A0A0L7KA08 S4PX45 A0A1B6DBJ7 A0A088AN65 A0A1B0CEA7 W8B448 A0A2A4J9C6 A0A2P8Z3D4 A0A1B6MA73 A0A2W1BTC8 A0A1B6I0C2 A0A1B6JJ87 B4KUP5 A0A0A1XLT4 A0A1J1I4U7 A0A336MN48 E9GQD7 B4LHS3 B4PCD0 D3TLW3 A0A1B0DAP6 A0A1L8DPQ1 B4QKZ0 A0A0L0BPC5 A0A1A9ZMR2 A0A1L8DXL4 U5EP87 B4HIX6 Q95TA4 A0A1W4VNI4 A0A0P5FWM6 A0A0P6AGE2 A0A0P6JJU3 Q9VSC3 Q24485 Q5U199 A0A336M662 B3NF74 A0A0P5M3G1 A0A2J7PTZ5 A0A067RJE8 A0A0P5LAE3 A0A0P5GJK9 A0A034VRC3 A0A0N8CV52 A0A0M4EJW8 B4J107 A0A0N1ITF6 A0A0K8WAA3 A0A0P5C0W7 A0A0N8A6Q3 A0A1A9WCE6 A0A1A9UVT1 Q29EP9 A0A1B6FQF4 A0A1Q3FNT2 A0A0L7RDF1 F4W570 B4H2B9 B3M5P6 Q1HQZ4 A0A1W4X0F2 A0A3B0K2R9 A0A158NYY0 A0A023ENC4 A0A1B0AQ50 A0A2A3EE73 A0A1W6EWB1 A0A1A9XTW2 B4N5I7 A0A2M3Z635 A0A1S3IDK3 A0A2M3Z6X1 B0WUA5 T1PE88 A0A1I8M8I6 A0A2M4AIE7 A0A2M4AI59 T1GHR4 W5J922 A0A195D0W7 A0A0T6AVS1 A0A0P4VI06 A0A0A9Z961 A0A146KWQ2 A0A0V0G625 E9IUX5 T1IEB5 A0A224XGW2 A0A232EJ98 U4UBW7
Pubmed
19121390
28756777
26354079
22651552
22118469
26227816
+ More
23622113 24495485 29403074 17994087 25830018 21292972 17550304 20353571 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7607542 24845553 25348373 15632085 21719571 17204158 17510324 21347285 24945155 25315136 20920257 23761445 27129103 25401762 26823975 21282665 28648823 23537049
23622113 24495485 29403074 17994087 25830018 21292972 17550304 20353571 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7607542 24845553 25348373 15632085 21719571 17204158 17510324 21347285 24945155 25315136 20920257 23761445 27129103 25401762 26823975 21282665 28648823 23537049
EMBL
BABH01005299
ODYU01003339
SOQ41962.1
KZ150015
PZC74985.1
NWSH01002283
+ More
PCG68687.1 RSAL01000028 RVE51874.1 KQ461194 KPJ06894.1 KQ459603 KPI92943.1 AK403899 BAM20015.1 AGBW02012613 OWR44721.1 JTDY01010212 KOB59940.1 GAIX01005678 JAA86882.1 GEDC01014288 JAS23010.1 AJWK01008710 GAMC01014717 JAB91838.1 PCG68685.1 PYGN01000215 PSN51011.1 GEBQ01031434 GEBQ01007156 JAT08543.1 JAT32821.1 PZC74983.1 GECU01037365 GECU01027343 GECU01020107 GECU01012462 JAS70341.1 JAS80363.1 JAS87599.1 JAS95244.1 GECU01033845 GECU01032693 GECU01028046 GECU01008507 JAS73861.1 JAS75013.1 JAS79660.1 JAS99199.1 CH933809 EDW18273.1 GBXI01002375 JAD11917.1 CVRI01000037 CRK93401.1 UFQS01000824 UFQT01000824 SSX07065.1 SSX27408.1 GL732558 EFX78360.1 CH940647 EDW70648.1 CM000159 EDW93815.1 CCAG010021538 EZ422415 ADD18691.1 AJVK01004585 GFDF01005665 JAV08419.1 CM000363 CM002912 EDX09623.1 KMY98252.1 JRES01001665 JRES01000968 KNC21079.1 KNC26671.1 GFDF01002935 JAV11149.1 GANO01000265 JAB59606.1 CH480815 EDW40630.1 AY060258 AAL25297.1 GDIQ01249372 JAK02353.1 GDIP01029745 JAM73970.1 GDIQ01005229 JAN89508.1 AE014296 AAF50502.1 X74932 CAA52884.1 BT015993 AAV36878.1 UFQS01000610 UFQT01000610 SSX05344.1 SSX25705.1 CH954178 EDV50416.1 GDIQ01161110 JAK90615.1 NEVH01021216 PNF19797.1 KK852649 KDR19500.1 GDIQ01182554 JAK69171.1 GDIQ01239580 JAK12145.1 GAKP01014851 GAKP01014849 GAKP01014847 JAC44103.1 GDIP01095831 JAM07884.1 CP012525 ALC43397.1 CH916366 EDV96862.1 KQ435824 KOX72128.1 GDHF01004191 JAI48123.1 GDIP01177114 JAJ46288.1 GDIP01177113 JAJ46289.1 CH379070 EAL30011.2 GECZ01017341 JAS52428.1 GFDL01005897 JAV29148.1 KQ414613 KOC69007.1 GL887605 EGI70666.1 CH479203 EDW30471.1 CH902618 EDV40680.2 DQ440300 CH477209 ABF18333.1 EAT47712.1 OUUW01000012 SPP86952.1 ADTU01004398 GAPW01002680 GEHC01000533 JAC10918.1 JAV47112.1 JXJN01001674 KZ288269 PBC30010.1 KY563593 ARK20002.1 CH964101 EDW79626.1 GGFM01003231 MBW23982.1 GGFM01003437 MBW24188.1 DS232103 EDS34877.1 KA647092 AFP61721.1 GGFK01007196 MBW40517.1 GGFK01006977 MBW40298.1 CAQQ02054978 ADMH02001841 ETN60942.1 KQ977004 KYN06507.1 LJIG01022683 KRT79267.1 GDKW01003212 JAI53383.1 GBHO01005259 GBRD01000269 JAG38345.1 JAG65552.1 GDHC01017715 JAQ00914.1 GECL01002622 JAP03502.1 GL766096 EFZ15615.1 ACPB03020873 GFTR01004689 JAW11737.1 NNAY01004054 OXU18440.1 KB632191 ERL89818.1
PCG68687.1 RSAL01000028 RVE51874.1 KQ461194 KPJ06894.1 KQ459603 KPI92943.1 AK403899 BAM20015.1 AGBW02012613 OWR44721.1 JTDY01010212 KOB59940.1 GAIX01005678 JAA86882.1 GEDC01014288 JAS23010.1 AJWK01008710 GAMC01014717 JAB91838.1 PCG68685.1 PYGN01000215 PSN51011.1 GEBQ01031434 GEBQ01007156 JAT08543.1 JAT32821.1 PZC74983.1 GECU01037365 GECU01027343 GECU01020107 GECU01012462 JAS70341.1 JAS80363.1 JAS87599.1 JAS95244.1 GECU01033845 GECU01032693 GECU01028046 GECU01008507 JAS73861.1 JAS75013.1 JAS79660.1 JAS99199.1 CH933809 EDW18273.1 GBXI01002375 JAD11917.1 CVRI01000037 CRK93401.1 UFQS01000824 UFQT01000824 SSX07065.1 SSX27408.1 GL732558 EFX78360.1 CH940647 EDW70648.1 CM000159 EDW93815.1 CCAG010021538 EZ422415 ADD18691.1 AJVK01004585 GFDF01005665 JAV08419.1 CM000363 CM002912 EDX09623.1 KMY98252.1 JRES01001665 JRES01000968 KNC21079.1 KNC26671.1 GFDF01002935 JAV11149.1 GANO01000265 JAB59606.1 CH480815 EDW40630.1 AY060258 AAL25297.1 GDIQ01249372 JAK02353.1 GDIP01029745 JAM73970.1 GDIQ01005229 JAN89508.1 AE014296 AAF50502.1 X74932 CAA52884.1 BT015993 AAV36878.1 UFQS01000610 UFQT01000610 SSX05344.1 SSX25705.1 CH954178 EDV50416.1 GDIQ01161110 JAK90615.1 NEVH01021216 PNF19797.1 KK852649 KDR19500.1 GDIQ01182554 JAK69171.1 GDIQ01239580 JAK12145.1 GAKP01014851 GAKP01014849 GAKP01014847 JAC44103.1 GDIP01095831 JAM07884.1 CP012525 ALC43397.1 CH916366 EDV96862.1 KQ435824 KOX72128.1 GDHF01004191 JAI48123.1 GDIP01177114 JAJ46288.1 GDIP01177113 JAJ46289.1 CH379070 EAL30011.2 GECZ01017341 JAS52428.1 GFDL01005897 JAV29148.1 KQ414613 KOC69007.1 GL887605 EGI70666.1 CH479203 EDW30471.1 CH902618 EDV40680.2 DQ440300 CH477209 ABF18333.1 EAT47712.1 OUUW01000012 SPP86952.1 ADTU01004398 GAPW01002680 GEHC01000533 JAC10918.1 JAV47112.1 JXJN01001674 KZ288269 PBC30010.1 KY563593 ARK20002.1 CH964101 EDW79626.1 GGFM01003231 MBW23982.1 GGFM01003437 MBW24188.1 DS232103 EDS34877.1 KA647092 AFP61721.1 GGFK01007196 MBW40517.1 GGFK01006977 MBW40298.1 CAQQ02054978 ADMH02001841 ETN60942.1 KQ977004 KYN06507.1 LJIG01022683 KRT79267.1 GDKW01003212 JAI53383.1 GBHO01005259 GBRD01000269 JAG38345.1 JAG65552.1 GDHC01017715 JAQ00914.1 GECL01002622 JAP03502.1 GL766096 EFZ15615.1 ACPB03020873 GFTR01004689 JAW11737.1 NNAY01004054 OXU18440.1 KB632191 ERL89818.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000005203 UP000092461 UP000245037 UP000009192 UP000183832 UP000000305 UP000008792 UP000002282 UP000092444 UP000092462 UP000000304 UP000037069 UP000092445 UP000001292 UP000192221 UP000000803 UP000008711 UP000235965 UP000027135 UP000092553 UP000001070 UP000053105 UP000091820 UP000078200 UP000001819 UP000053825 UP000007755 UP000008744 UP000007801 UP000008820 UP000192223 UP000268350 UP000005205 UP000092460 UP000242457 UP000092443 UP000007798 UP000085678 UP000002320 UP000095301 UP000015102 UP000000673 UP000078542 UP000015103 UP000215335 UP000030742
UP000037510 UP000005203 UP000092461 UP000245037 UP000009192 UP000183832 UP000000305 UP000008792 UP000002282 UP000092444 UP000092462 UP000000304 UP000037069 UP000092445 UP000001292 UP000192221 UP000000803 UP000008711 UP000235965 UP000027135 UP000092553 UP000001070 UP000053105 UP000091820 UP000078200 UP000001819 UP000053825 UP000007755 UP000008744 UP000007801 UP000008820 UP000192223 UP000268350 UP000005205 UP000092460 UP000242457 UP000092443 UP000007798 UP000085678 UP000002320 UP000095301 UP000015102 UP000000673 UP000078542 UP000015103 UP000215335 UP000030742
Interpro
IPR036430
RNase_T2-like_sf
+ More
IPR001568 RNase_T2-like
IPR033130 RNase_T2_His_AS_2
IPR033697 Ribonuclease_T2_eukaryotic
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR003593 AAA+_ATPase
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR027417 P-loop_NTPase
IPR018188 RNase_T2_His_AS_1
IPR013525 ABC_2_trans
IPR001568 RNase_T2-like
IPR033130 RNase_T2_His_AS_2
IPR033697 Ribonuclease_T2_eukaryotic
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR003593 AAA+_ATPase
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR027417 P-loop_NTPase
IPR018188 RNase_T2_His_AS_1
IPR013525 ABC_2_trans
Gene 3D
ProteinModelPortal
H9JLW8
A0A2H1VND0
A0A2W1BMY3
A0A2A4J987
A0A3S2LQR4
A0A194QNG1
+ More
A0A194PIR6 I4DQ25 A0A212ETF2 A0A0L7KA08 S4PX45 A0A1B6DBJ7 A0A088AN65 A0A1B0CEA7 W8B448 A0A2A4J9C6 A0A2P8Z3D4 A0A1B6MA73 A0A2W1BTC8 A0A1B6I0C2 A0A1B6JJ87 B4KUP5 A0A0A1XLT4 A0A1J1I4U7 A0A336MN48 E9GQD7 B4LHS3 B4PCD0 D3TLW3 A0A1B0DAP6 A0A1L8DPQ1 B4QKZ0 A0A0L0BPC5 A0A1A9ZMR2 A0A1L8DXL4 U5EP87 B4HIX6 Q95TA4 A0A1W4VNI4 A0A0P5FWM6 A0A0P6AGE2 A0A0P6JJU3 Q9VSC3 Q24485 Q5U199 A0A336M662 B3NF74 A0A0P5M3G1 A0A2J7PTZ5 A0A067RJE8 A0A0P5LAE3 A0A0P5GJK9 A0A034VRC3 A0A0N8CV52 A0A0M4EJW8 B4J107 A0A0N1ITF6 A0A0K8WAA3 A0A0P5C0W7 A0A0N8A6Q3 A0A1A9WCE6 A0A1A9UVT1 Q29EP9 A0A1B6FQF4 A0A1Q3FNT2 A0A0L7RDF1 F4W570 B4H2B9 B3M5P6 Q1HQZ4 A0A1W4X0F2 A0A3B0K2R9 A0A158NYY0 A0A023ENC4 A0A1B0AQ50 A0A2A3EE73 A0A1W6EWB1 A0A1A9XTW2 B4N5I7 A0A2M3Z635 A0A1S3IDK3 A0A2M3Z6X1 B0WUA5 T1PE88 A0A1I8M8I6 A0A2M4AIE7 A0A2M4AI59 T1GHR4 W5J922 A0A195D0W7 A0A0T6AVS1 A0A0P4VI06 A0A0A9Z961 A0A146KWQ2 A0A0V0G625 E9IUX5 T1IEB5 A0A224XGW2 A0A232EJ98 U4UBW7
A0A194PIR6 I4DQ25 A0A212ETF2 A0A0L7KA08 S4PX45 A0A1B6DBJ7 A0A088AN65 A0A1B0CEA7 W8B448 A0A2A4J9C6 A0A2P8Z3D4 A0A1B6MA73 A0A2W1BTC8 A0A1B6I0C2 A0A1B6JJ87 B4KUP5 A0A0A1XLT4 A0A1J1I4U7 A0A336MN48 E9GQD7 B4LHS3 B4PCD0 D3TLW3 A0A1B0DAP6 A0A1L8DPQ1 B4QKZ0 A0A0L0BPC5 A0A1A9ZMR2 A0A1L8DXL4 U5EP87 B4HIX6 Q95TA4 A0A1W4VNI4 A0A0P5FWM6 A0A0P6AGE2 A0A0P6JJU3 Q9VSC3 Q24485 Q5U199 A0A336M662 B3NF74 A0A0P5M3G1 A0A2J7PTZ5 A0A067RJE8 A0A0P5LAE3 A0A0P5GJK9 A0A034VRC3 A0A0N8CV52 A0A0M4EJW8 B4J107 A0A0N1ITF6 A0A0K8WAA3 A0A0P5C0W7 A0A0N8A6Q3 A0A1A9WCE6 A0A1A9UVT1 Q29EP9 A0A1B6FQF4 A0A1Q3FNT2 A0A0L7RDF1 F4W570 B4H2B9 B3M5P6 Q1HQZ4 A0A1W4X0F2 A0A3B0K2R9 A0A158NYY0 A0A023ENC4 A0A1B0AQ50 A0A2A3EE73 A0A1W6EWB1 A0A1A9XTW2 B4N5I7 A0A2M3Z635 A0A1S3IDK3 A0A2M3Z6X1 B0WUA5 T1PE88 A0A1I8M8I6 A0A2M4AIE7 A0A2M4AI59 T1GHR4 W5J922 A0A195D0W7 A0A0T6AVS1 A0A0P4VI06 A0A0A9Z961 A0A146KWQ2 A0A0V0G625 E9IUX5 T1IEB5 A0A224XGW2 A0A232EJ98 U4UBW7
PDB
3T0O
E-value=5.98448e-26,
Score=290
Ontologies
GO
PANTHER
Topology
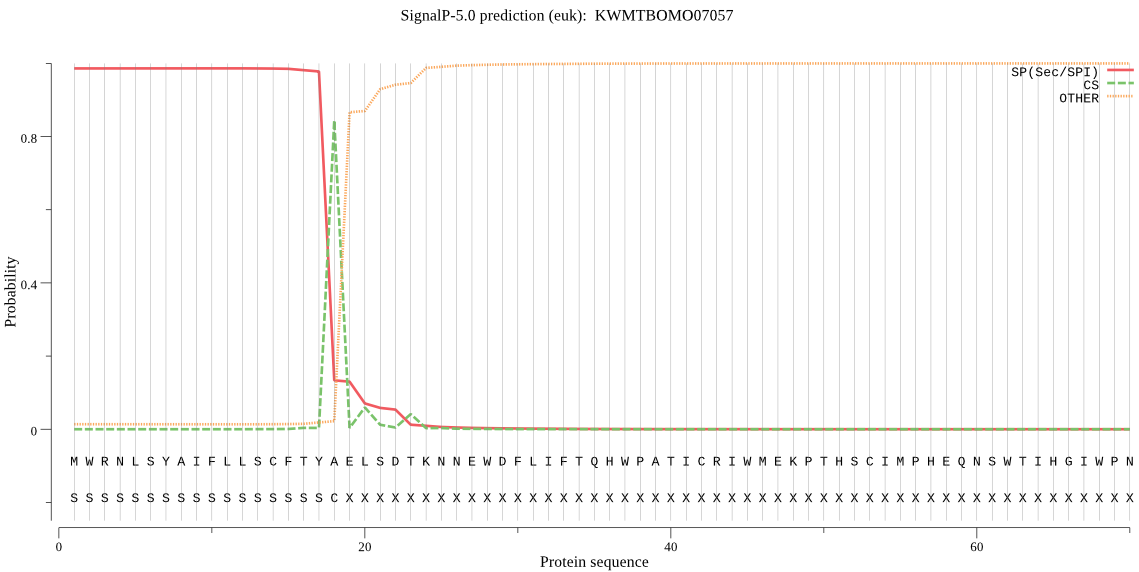
SignalP
Position: 1 - 18,
Likelihood: 0.986120
Length:
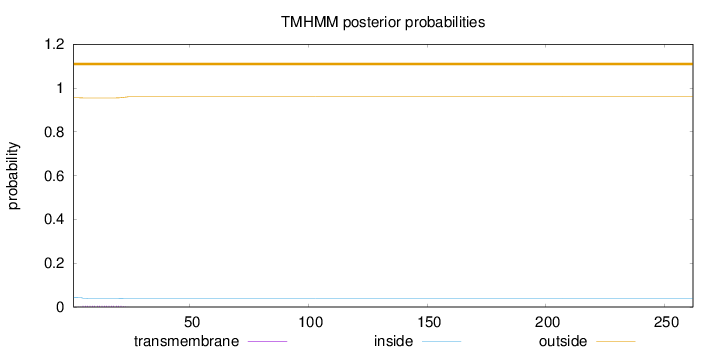
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10163
Exp number, first 60 AAs:
0.10163
Total prob of N-in:
0.04444
outside
1 - 262
Population Genetic Test Statistics
Pi
177.807095
Theta
143.53233
Tajima's D
0.35451
CLR
0.071134
CSRT
0.475126243687816
Interpretation
Uncertain
