Gene
KWMTBOMO07050 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010515
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.386
Sequence
CDS
ATGGACTTTTTCAGCAATTTCATTGCGTCGGCTGAGGTTATGTCGTTAAAACCATTCACGGAGCTATTGCAAGCCGTTTTTGCTGTACTGGCAACATTGAACTTGAAGGTAGGAGGGTATCCGGAGCAACATGTCTTACAAAATAATACATCTTACGATATAATTATTGTCGGCGCTGGTTCATCTGGTTGCGTCCTGGCCAACCGACTCTCTGAGATCTCAAAGTGGACAGTGCTACTACTGGAGGCCGGAGAAGATCCTTCCATTGAATCCACGATACCGGGATTATTTCCATTTGTTGATTACTCGTACGAAGATTGGAACTACAGAACCGTTAAGAACAATTATTCATGTCAAGCGCATACAGAAAAAAATATTCATTTGACGAGAGGCAAAATGGTTGGTGGCTGTAGTGGATCAAACTATATGTATTACGCCCGAGGGAACAGACATGATTATGACTCTTGGGAAGGAATGGGCAACAAAGGGTGGGGCTACAAATCAGTACTTCCATATTTTATTAAAAGTGAACGACTTGAAGATGACACAATATTGAAGAGCCCCTCTGGATCTTTCCACGGGACAGAAGGTTACCTCGGTGTTACGAAACCAGACTGGGGGGATAGAGTAGATGACTATTTTACAGCATTCAAACAAAATGAACGTAAAGTTATATTGGACCAGAACGGACCGGAGCAGATGGGTTACGCACTACCGACATTCACTGTTGCTGATAATGTTCGCCAGAGCACAGGCTACGCTTTCTTGAGACCCATAAAAAATCGTAAAAATTTGTTTCTTTCAAGAAAGTCTTTCGTCAGGAAAGTTTTATTTAATGATGCAAAAGAAGCAGAAGCTGTAGAAGTCATGCAAAATGGTAAAATACTGATAATTAAAGCAAGACAAGAAATAATAATATCCGCTGGTGCAATAAACAGCCCGCAGCTATTGATGCTGTCAGGAATCGGCCCAAAAGAGCATCTAACTGAGATGGCCATTCCAGTTATCAGTGAATCACCAAACGTCGGAAAAAACTTACAAGACCATGCCACTGTTCCACTGATCATCACTGAAAATATAAGCAAGGTCACTGTTGTTCAAAATGTTGAAACATTTACCAATTTAAATGAGATGCCCGCACCGTGTCTTATGGGATTCGCTGCCGTCAATAAAACGCAAACTATACCAGACTACCAAGTCAAATTTTTCGTATTTCCCGTTGGTACTATAATTCCTGTTTGGATTTGTAACTTTGTATTCGGTTTGGCTGACAGCATTTGCGTGTCTATGGCGAAGGCAGCACAGAAAAAAGGTGTTATTTTAGCTCTTGTAGTCCTTTTGTATCCAAAATCAACTGGTAAAGTTGTTCTTAAAAGCAAAAACCCTGAAGAACCTCCCTTAATTTACACAGGCTATTACTCTCAAGATGAAGATTTAGAAAATCATGCAAAATACATAGAAGACTTCTTGACTGTGCTCGATACAGAACACATGAGAAAAACTCACGCGCAACCTCTGAACCTTAATGTAAAACAATGCAAACATCACAAGTTTAATACCAGAAAATATTGGAAGTGTTTTGCTTTGAATGTAGTTACAACTCTCTGGCATCCTGTCGCTACTTGCGCTATGGGTCCTGAAGGAGTTGGGGTAATTGATGACAAACTGAGAGTACATGGTGTGAAGAAATTAAGAGTCATTGATGCCAGTTCGATGCCGAAGATAGTCGGTGGGAATACAAATGCTCCTACCATCATGATTGCTGATAAAGGAGCTGATATCATCAAAGATGATCACGGCTTAATATGA
Protein
MDFFSNFIASAEVMSLKPFTELLQAVFAVLATLNLKVGGYPEQHVLQNNTSYDIIIVGAGSSGCVLANRLSEISKWTVLLLEAGEDPSIESTIPGLFPFVDYSYEDWNYRTVKNNYSCQAHTEKNIHLTRGKMVGGCSGSNYMYYARGNRHDYDSWEGMGNKGWGYKSVLPYFIKSERLEDDTILKSPSGSFHGTEGYLGVTKPDWGDRVDDYFTAFKQNERKVILDQNGPEQMGYALPTFTVADNVRQSTGYAFLRPIKNRKNLFLSRKSFVRKVLFNDAKEAEAVEVMQNGKILIIKARQEIIISAGAINSPQLLMLSGIGPKEHLTEMAIPVISESPNVGKNLQDHATVPLIITENISKVTVVQNVETFTNLNEMPAPCLMGFAAVNKTQTIPDYQVKFFVFPVGTIIPVWICNFVFGLADSICVSMAKAAQKKGVILALVVLLYPKSTGKVVLKSKNPEEPPLIYTGYYSQDEDLENHAKYIEDFLTVLDTEHMRKTHAQPLNLNVKQCKHHKFNTRKYWKCFALNVVTTLWHPVATCAMGPEGVGVIDDKLRVHGVKKLRVIDASSMPKIVGGNTNAPTIMIADKGADIIKDDHGLI
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JLW3
H9JLW4
A0A0L7LUK3
A0A2A4IZE5
A0A2W1BIV7
A0A2H1VYA6
+ More
H9JLP6 A0A2A4K9G4 A0A2H1V0S8 A0A2W1B6G7 A0A194QNE6 A0A194PPA0 H9JLQ9 A0A0L7LQD8 A0A0L7L6H5 A0A2W1BQ09 A0A3S2LRK7 A0A3S2N607 A0A212EII3 H9JRF5 A0A3S2M5K0 A0A0L7LP50 A0A0L7KKV3 A0A0L7LL99 A0A212F6I8 A0A0L7KZE8 A0A2W1BRA5 D0ABA6 A0A2A4JWF9 A0A0N1IPT1 A0A0N1PIV1 A0A2H1VCU6 A0A2H1VB55 A0A0N0PDR9 A0A2A4JYC1 A0A0L7LPB4 A0A212F6I0 A0A194QGE4 A0A212FMS7 A0A2H1VCV1 A0A2H1WHG3 A0A2H1W336 A0A194QAM1 A0A2A4IZ69 A0A2H1WPH8 A0A194QAR0 A0A194R898 A0A2W1BCG7 A0A2A4K6C3 E2ASE8 A0A2W1BIM1 A0A212F947 A0A2H1WZ72 A0A154PBS2 A0A2J7PXQ0 A0A194PXZ3 A0A2A4JF29 A0A232EPA2 A0A0L7RJZ7 A0A2H1W7M4 K7J280 A0A2A4K5W1 A0A0N1I9P3 H9JI92 A0A310SSQ6 E9IKG4 A0A1B6G5I8 A0A2A4J139 A0A1B6KBZ3 A0A1B6IAY6 A0A158NUR1 A0A1L8E0M0 A0A1L8E037 A0A2W1BM44 E2AAW1 A0A2K8FTP1 A0A088AJV0 A0A151I6J0 A0A0C9RDB5 A0A0N0U438 A0A151K0G8 A0A1S6J0X2 A0A1B0GIG3 A0A2H1VP10 A0A1B6EYD4 A0A1B0GMI8 A0A3L8D7B4 A0A2W1BU23 A0A1B0DMM3 A0A2J7Q2I0 A0A2H1W3T3 A0A2J7Q2J7 A0A2H1VDD9 A0A291P0W5 A0A2W1BWE7 E9JDH0 A0A023EVK5 A0A2R7W646 A0A2H1VDA1 A0A195BBF9
H9JLP6 A0A2A4K9G4 A0A2H1V0S8 A0A2W1B6G7 A0A194QNE6 A0A194PPA0 H9JLQ9 A0A0L7LQD8 A0A0L7L6H5 A0A2W1BQ09 A0A3S2LRK7 A0A3S2N607 A0A212EII3 H9JRF5 A0A3S2M5K0 A0A0L7LP50 A0A0L7KKV3 A0A0L7LL99 A0A212F6I8 A0A0L7KZE8 A0A2W1BRA5 D0ABA6 A0A2A4JWF9 A0A0N1IPT1 A0A0N1PIV1 A0A2H1VCU6 A0A2H1VB55 A0A0N0PDR9 A0A2A4JYC1 A0A0L7LPB4 A0A212F6I0 A0A194QGE4 A0A212FMS7 A0A2H1VCV1 A0A2H1WHG3 A0A2H1W336 A0A194QAM1 A0A2A4IZ69 A0A2H1WPH8 A0A194QAR0 A0A194R898 A0A2W1BCG7 A0A2A4K6C3 E2ASE8 A0A2W1BIM1 A0A212F947 A0A2H1WZ72 A0A154PBS2 A0A2J7PXQ0 A0A194PXZ3 A0A2A4JF29 A0A232EPA2 A0A0L7RJZ7 A0A2H1W7M4 K7J280 A0A2A4K5W1 A0A0N1I9P3 H9JI92 A0A310SSQ6 E9IKG4 A0A1B6G5I8 A0A2A4J139 A0A1B6KBZ3 A0A1B6IAY6 A0A158NUR1 A0A1L8E0M0 A0A1L8E037 A0A2W1BM44 E2AAW1 A0A2K8FTP1 A0A088AJV0 A0A151I6J0 A0A0C9RDB5 A0A0N0U438 A0A151K0G8 A0A1S6J0X2 A0A1B0GIG3 A0A2H1VP10 A0A1B6EYD4 A0A1B0GMI8 A0A3L8D7B4 A0A2W1BU23 A0A1B0DMM3 A0A2J7Q2I0 A0A2H1W3T3 A0A2J7Q2J7 A0A2H1VDD9 A0A291P0W5 A0A2W1BWE7 E9JDH0 A0A023EVK5 A0A2R7W646 A0A2H1VDA1 A0A195BBF9
Pubmed
EMBL
BABH01005279
BABH01005283
JTDY01000066
KOB79079.1
NWSH01004349
PCG65195.1
+ More
KZ150015 PZC74989.1 ODYU01005196 SOQ45815.1 BABH01005285 NWSH01000039 PCG80370.1 ODYU01000155 SOQ34448.1 KZ150274 PZC71662.1 KQ461194 KPJ06879.1 KQ459603 KPI92960.1 BABH01005224 BABH01005225 BABH01005226 JTDY01000335 KOB77647.1 JTDY01002689 KOB70914.1 KZ150085 PZC73783.1 RSAL01000341 RVE42399.1 RVE42398.1 AGBW02014643 OWR41285.1 BABH01016356 RSAL01000028 RVE51877.1 JTDY01000479 KOB76996.1 JTDY01009199 KOB63957.1 JTDY01000758 KOB75991.1 AGBW02010007 OWR49347.1 JTDY01004150 KOB68525.1 KZ149986 PZC75677.1 FP102341 CBH09301.1 NWSH01000445 PCG76355.1 KQ460044 KPJ18289.1 KPJ18290.1 ODYU01001603 SOQ38054.1 ODYU01001607 SOQ38065.1 KPJ18288.1 NWSH01000427 PCG76483.1 JTDY01000418 KOB77282.1 OWR49345.1 KQ459232 KPJ02521.1 AGBW02007651 OWR55045.1 SOQ38064.1 ODYU01008704 SOQ52509.1 ODYU01006017 SOQ47491.1 KPJ02522.1 NWSH01004322 PCG65227.1 ODYU01010073 SOQ54917.1 KPJ02519.1 KQ460627 KPJ13480.1 KZ150303 PZC71524.1 NWSH01000095 PCG79619.1 GL442298 EFN63614.1 PZC71523.1 AGBW02009643 OWR50262.1 ODYU01012146 SOQ58297.1 KQ434869 KZC09325.1 NEVH01020853 PNF21103.1 KQ459585 KPI98211.1 NWSH01001625 PCG70657.1 NNAY01002979 OXU20190.1 KQ414579 KOC71128.1 ODYU01006844 SOQ49058.1 AAZX01001588 PCG79621.1 KPJ18287.1 BABH01022249 KQ759784 OAD62857.1 GL763984 EFZ18942.1 GECZ01024966 GECZ01012070 JAS44803.1 JAS57699.1 NWSH01004424 PCG65132.1 GEBQ01031263 GEBQ01012250 JAT08714.1 JAT27727.1 GECU01023601 JAS84105.1 ADTU01026684 ADTU01026685 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 PZC73780.1 GL438191 EFN69424.1 KY618826 AQW43012.1 KQ978473 KYM93711.1 GBYB01011122 JAG80889.1 KQ435846 KOX71152.1 KQ981296 KYN43079.1 KU764422 AQS60669.1 AJWK01014080 ODYU01003587 SOQ42538.1 GECZ01026787 JAS42982.1 AJVK01024561 AJVK01024562 QOIP01000012 RLU16179.1 KZ149963 PZC76280.1 AJVK01016891 NEVH01019370 PNF22796.1 SOQ47492.1 PNF22797.1 ODYU01001944 SOQ38845.1 MF687606 ATJ44532.1 KZ149916 PZC77944.1 GL771866 EFZ09157.1 GAPW01000526 JAC13072.1 KK854321 PTY14631.1 ODYU01001935 SOQ38829.1 KQ976532 KYM81562.1
KZ150015 PZC74989.1 ODYU01005196 SOQ45815.1 BABH01005285 NWSH01000039 PCG80370.1 ODYU01000155 SOQ34448.1 KZ150274 PZC71662.1 KQ461194 KPJ06879.1 KQ459603 KPI92960.1 BABH01005224 BABH01005225 BABH01005226 JTDY01000335 KOB77647.1 JTDY01002689 KOB70914.1 KZ150085 PZC73783.1 RSAL01000341 RVE42399.1 RVE42398.1 AGBW02014643 OWR41285.1 BABH01016356 RSAL01000028 RVE51877.1 JTDY01000479 KOB76996.1 JTDY01009199 KOB63957.1 JTDY01000758 KOB75991.1 AGBW02010007 OWR49347.1 JTDY01004150 KOB68525.1 KZ149986 PZC75677.1 FP102341 CBH09301.1 NWSH01000445 PCG76355.1 KQ460044 KPJ18289.1 KPJ18290.1 ODYU01001603 SOQ38054.1 ODYU01001607 SOQ38065.1 KPJ18288.1 NWSH01000427 PCG76483.1 JTDY01000418 KOB77282.1 OWR49345.1 KQ459232 KPJ02521.1 AGBW02007651 OWR55045.1 SOQ38064.1 ODYU01008704 SOQ52509.1 ODYU01006017 SOQ47491.1 KPJ02522.1 NWSH01004322 PCG65227.1 ODYU01010073 SOQ54917.1 KPJ02519.1 KQ460627 KPJ13480.1 KZ150303 PZC71524.1 NWSH01000095 PCG79619.1 GL442298 EFN63614.1 PZC71523.1 AGBW02009643 OWR50262.1 ODYU01012146 SOQ58297.1 KQ434869 KZC09325.1 NEVH01020853 PNF21103.1 KQ459585 KPI98211.1 NWSH01001625 PCG70657.1 NNAY01002979 OXU20190.1 KQ414579 KOC71128.1 ODYU01006844 SOQ49058.1 AAZX01001588 PCG79621.1 KPJ18287.1 BABH01022249 KQ759784 OAD62857.1 GL763984 EFZ18942.1 GECZ01024966 GECZ01012070 JAS44803.1 JAS57699.1 NWSH01004424 PCG65132.1 GEBQ01031263 GEBQ01012250 JAT08714.1 JAT27727.1 GECU01023601 JAS84105.1 ADTU01026684 ADTU01026685 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 PZC73780.1 GL438191 EFN69424.1 KY618826 AQW43012.1 KQ978473 KYM93711.1 GBYB01011122 JAG80889.1 KQ435846 KOX71152.1 KQ981296 KYN43079.1 KU764422 AQS60669.1 AJWK01014080 ODYU01003587 SOQ42538.1 GECZ01026787 JAS42982.1 AJVK01024561 AJVK01024562 QOIP01000012 RLU16179.1 KZ149963 PZC76280.1 AJVK01016891 NEVH01019370 PNF22796.1 SOQ47492.1 PNF22797.1 ODYU01001944 SOQ38845.1 MF687606 ATJ44532.1 KZ149916 PZC77944.1 GL771866 EFZ09157.1 GAPW01000526 JAC13072.1 KK854321 PTY14631.1 ODYU01001935 SOQ38829.1 KQ976532 KYM81562.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JLW3
H9JLW4
A0A0L7LUK3
A0A2A4IZE5
A0A2W1BIV7
A0A2H1VYA6
+ More
H9JLP6 A0A2A4K9G4 A0A2H1V0S8 A0A2W1B6G7 A0A194QNE6 A0A194PPA0 H9JLQ9 A0A0L7LQD8 A0A0L7L6H5 A0A2W1BQ09 A0A3S2LRK7 A0A3S2N607 A0A212EII3 H9JRF5 A0A3S2M5K0 A0A0L7LP50 A0A0L7KKV3 A0A0L7LL99 A0A212F6I8 A0A0L7KZE8 A0A2W1BRA5 D0ABA6 A0A2A4JWF9 A0A0N1IPT1 A0A0N1PIV1 A0A2H1VCU6 A0A2H1VB55 A0A0N0PDR9 A0A2A4JYC1 A0A0L7LPB4 A0A212F6I0 A0A194QGE4 A0A212FMS7 A0A2H1VCV1 A0A2H1WHG3 A0A2H1W336 A0A194QAM1 A0A2A4IZ69 A0A2H1WPH8 A0A194QAR0 A0A194R898 A0A2W1BCG7 A0A2A4K6C3 E2ASE8 A0A2W1BIM1 A0A212F947 A0A2H1WZ72 A0A154PBS2 A0A2J7PXQ0 A0A194PXZ3 A0A2A4JF29 A0A232EPA2 A0A0L7RJZ7 A0A2H1W7M4 K7J280 A0A2A4K5W1 A0A0N1I9P3 H9JI92 A0A310SSQ6 E9IKG4 A0A1B6G5I8 A0A2A4J139 A0A1B6KBZ3 A0A1B6IAY6 A0A158NUR1 A0A1L8E0M0 A0A1L8E037 A0A2W1BM44 E2AAW1 A0A2K8FTP1 A0A088AJV0 A0A151I6J0 A0A0C9RDB5 A0A0N0U438 A0A151K0G8 A0A1S6J0X2 A0A1B0GIG3 A0A2H1VP10 A0A1B6EYD4 A0A1B0GMI8 A0A3L8D7B4 A0A2W1BU23 A0A1B0DMM3 A0A2J7Q2I0 A0A2H1W3T3 A0A2J7Q2J7 A0A2H1VDD9 A0A291P0W5 A0A2W1BWE7 E9JDH0 A0A023EVK5 A0A2R7W646 A0A2H1VDA1 A0A195BBF9
H9JLP6 A0A2A4K9G4 A0A2H1V0S8 A0A2W1B6G7 A0A194QNE6 A0A194PPA0 H9JLQ9 A0A0L7LQD8 A0A0L7L6H5 A0A2W1BQ09 A0A3S2LRK7 A0A3S2N607 A0A212EII3 H9JRF5 A0A3S2M5K0 A0A0L7LP50 A0A0L7KKV3 A0A0L7LL99 A0A212F6I8 A0A0L7KZE8 A0A2W1BRA5 D0ABA6 A0A2A4JWF9 A0A0N1IPT1 A0A0N1PIV1 A0A2H1VCU6 A0A2H1VB55 A0A0N0PDR9 A0A2A4JYC1 A0A0L7LPB4 A0A212F6I0 A0A194QGE4 A0A212FMS7 A0A2H1VCV1 A0A2H1WHG3 A0A2H1W336 A0A194QAM1 A0A2A4IZ69 A0A2H1WPH8 A0A194QAR0 A0A194R898 A0A2W1BCG7 A0A2A4K6C3 E2ASE8 A0A2W1BIM1 A0A212F947 A0A2H1WZ72 A0A154PBS2 A0A2J7PXQ0 A0A194PXZ3 A0A2A4JF29 A0A232EPA2 A0A0L7RJZ7 A0A2H1W7M4 K7J280 A0A2A4K5W1 A0A0N1I9P3 H9JI92 A0A310SSQ6 E9IKG4 A0A1B6G5I8 A0A2A4J139 A0A1B6KBZ3 A0A1B6IAY6 A0A158NUR1 A0A1L8E0M0 A0A1L8E037 A0A2W1BM44 E2AAW1 A0A2K8FTP1 A0A088AJV0 A0A151I6J0 A0A0C9RDB5 A0A0N0U438 A0A151K0G8 A0A1S6J0X2 A0A1B0GIG3 A0A2H1VP10 A0A1B6EYD4 A0A1B0GMI8 A0A3L8D7B4 A0A2W1BU23 A0A1B0DMM3 A0A2J7Q2I0 A0A2H1W3T3 A0A2J7Q2J7 A0A2H1VDD9 A0A291P0W5 A0A2W1BWE7 E9JDH0 A0A023EVK5 A0A2R7W646 A0A2H1VDA1 A0A195BBF9
PDB
5NCC
E-value=1.65748e-48,
Score=488
Ontologies
GO
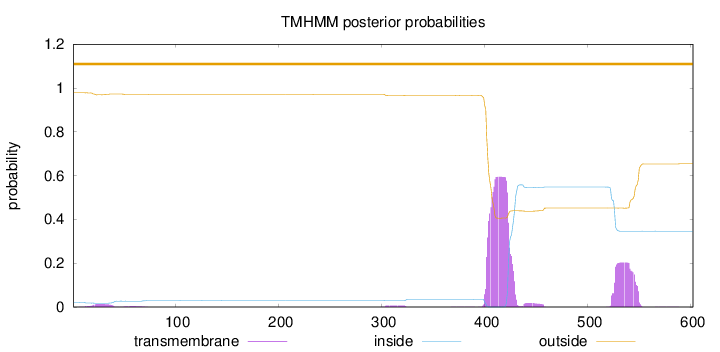
Topology
Length:
602
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
18.02957
Exp number, first 60 AAs:
0.31676
Total prob of N-in:
0.02166
outside
1 - 602
Population Genetic Test Statistics
Pi
151.628109
Theta
147.845541
Tajima's D
-0.838837
CLR
0.85949
CSRT
0.171891405429728
Interpretation
Uncertain
