Pre Gene Modal
BGIBMGA010449
Annotation
PREDICTED:_TBC1_domain_family_member_15_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.004
Sequence
CDS
ATGTCTACGAGTTCGGGAGATGAATTACACGACCAGTACAAAGACTTGTTCACGCAAGACGGGGTACTTCTAAAGACCAGTGTCGGTCGGCCGGTCTCCGATTTCAACTCAATCGGGATGCTCTGCATTGTACAGAACGCCGACGGAACTAAATGCATAGAATGGAGACCGAACGACTTGATCACCATCGATTCCGACACTCAAGATCAAGAATGGGCCGTTGTTAATACCGTTGGTCGCAGACAACGGACCCTAAGCGGTAACATAACATCGGATTACGCGAATCTTCGGGCGCGCATCATAAAAATACCCCTAGACGATTTAAAAACGTTTCGCATCACCCGTAACAACCAACAAATGCAGTTCTCGACGAAACCGGGAGTCTGGCAGAATACGTTCTATTTCCAACATGGAAACGCCGAGATATTCGTGGCCTATCTAAAGAATCACCTTAAGACCGCTAAAACAAGACACGACAGGAACACGTACATGGTGGTCGAACCTAACGACGAGTCCCAGGTCTTGAATAAATCGTTTGCCGAATTAGATATATTTACGGAGAATACGACTGATGTCGTGTGGAATTTGGTTTCTAATTTCAAACAGAGGCCTTACGAAACTACCTTAGAAGCGTTTTCAAAGCTAACGGATATTGTCTACTACGGCAATGAGGATGTAAACGGGAAACGAGACGTTTCAGAGGAGGTCGCCGACTTGTTGACTAAAAGCATGAGCGCTCTCGAGGACACGACCTCGGTCACCAGGAGCGACGAGTACGAGGTCATCAGCGTGAAGCCAACGTTGCCTCCCCGGCCGTGTATACCCAGAGGTTCGCCACTGTCCACTGAAAAATGGGACGCCTTGCAAGACGCGGACGGTAGGATCGTGGAGGTGGAGGGAGTCAAGCAACTCATATTCAGGGGAGGTGTTGCACATTCTATACGCAGCACCGTATGGAAGTATCTCTTGGACTATTTCCCCTGGAAGATGACGCATGCAGAATTAAAAACGTTACAGAAGAGAAAAACCGAGGAATACTTCTCTATGAAACTCCAATGGAGGTCTGTGACCGAGGGGCAGGAGAATAGGTTCTCTGAGTATAGAGACAGGAAGAGTTTGGTCGAGAAGGATGTGAACAGAACGGACAGGACTCACCCGTTTTTCGCGGGGGACAACAACCCCCACTTGGCCGTGCTCCAGGACATCCTGATGACCTACGTCATGTACAACTTCGATCTCGGGTACGTGCAGGGAATGAGCGATATATTGGCGCCTCTGCTGCTGCTGTTAGGAAACGAAGTGGACAGTTTCTGGTGCTTCGTCGGCTTCATGGAGAAAATTGCTTCGAATTTCGACATGGACCAGGCCGGAATGAAGCACCAGCTCTTGCAGCTCCAGCAGCTGCTGATGTTCGCTAGCCCGGAGCTGGCCCAGCACCTGTCACAAAAGGACTCCGGGAACATGTACTTTTGCTTCCGCTGGCTGCTCGTATGGTTCAAAAGGGAATTCTCACACATCGATATCATGAGGCTGTGGGAAGTATTGTGGACGGGGTTACCATGTGCAAATTTTCATTTGTTGATATGCGTTGCCATATTAGATGCAGAAAAAGAAGTTATTATTAGTCGAGATTACGGTTTCACTGAAATATTAAAGCATGTGAACGATCTATCGATGACTTTGGACGTTGACAGGGTACTAAGTTCAGCCGAAGGCATATATCACCAGATAGTTTCAGCCCCGCATTTGACAGATCAAATCAGGATAGTCTTAGGAATCGAGCCATTGAATCTGTCAATGTCAACAAATTCCGACCCTGAGCCCTCTACCTCTACTCTACCCTCTACCAAACCAAGTCAAGTTGAGCTAAAGGAGGGCAACGGTAATACCATTTTTGGTCAAGAAATGGACGAAGATGTAGTCGTATATAATGGATTACGAATAACAATAGTTACGAAAACTGTATGGGCTCTACTCGCCTGA
Protein
MSTSSGDELHDQYKDLFTQDGVLLKTSVGRPVSDFNSIGMLCIVQNADGTKCIEWRPNDLITIDSDTQDQEWAVVNTVGRRQRTLSGNITSDYANLRARIIKIPLDDLKTFRITRNNQQMQFSTKPGVWQNTFYFQHGNAEIFVAYLKNHLKTAKTRHDRNTYMVVEPNDESQVLNKSFAELDIFTENTTDVVWNLVSNFKQRPYETTLEAFSKLTDIVYYGNEDVNGKRDVSEEVADLLTKSMSALEDTTSVTRSDEYEVISVKPTLPPRPCIPRGSPLSTEKWDALQDADGRIVEVEGVKQLIFRGGVAHSIRSTVWKYLLDYFPWKMTHAELKTLQKRKTEEYFSMKLQWRSVTEGQENRFSEYRDRKSLVEKDVNRTDRTHPFFAGDNNPHLAVLQDILMTYVMYNFDLGYVQGMSDILAPLLLLLGNEVDSFWCFVGFMEKIASNFDMDQAGMKHQLLQLQQLLMFASPELAQHLSQKDSGNMYFCFRWLLVWFKREFSHIDIMRLWEVLWTGLPCANFHLLICVAILDAEKEVIISRDYGFTEILKHVNDLSMTLDVDRVLSSAEGIYHQIVSAPHLTDQIRIVLGIEPLNLSMSTNSDPEPSTSTLPSTKPSQVELKEGNGNTIFGQEMDEDVVVYNGLRITIVTKTVWALLA
Summary
Uniprot
H9JLP7
A0A2W1BL30
A0A2H1W6T5
A0A1E1WCN4
A0A1E1W2B7
A0A194PHQ3
+ More
A0A212FIC0 A0A1Y1KQT1 A0A2J7RC25 A0A151WQT4 E9IQK4 A0A151J196 A0A158NYX6 A0A195BVF4 A0A195FT32 A0A154PAU5 D2A2J8 A0A1W4X1R8 A0A026WDF5 A0A3L8DWS5 A0A067RD98 A0A2J7RC26 A0A088AMZ8 A0A2A3EE34 V5GV59 A0A0L7RDW4 A0A1W4WS45 A0A195D0S0 A0A1B6CAJ1 A0A0M8ZYD8 A0A1B6G5P0 A0A1B6E0S5 A0A1B6MVH7 A0A1B6L3E5 K7J9T2 A0A232EVQ4 A0A2J7RC34 A0A0J7P142 F4W5H6 E2C0H0 A0A1B6GMV3 A0A224X940 A0A069DVG0 A0A0P4VKX6 A0A0V0G4Y8 A0A023F1U4 T1HPL0 A0A1B0DJW6 A0A1L8DLW6 A0A210Q8D9 A0A034WN52 A0A1S3JNQ6 K1PL63 A0A1A9VDR9 A0A0K8TWX7 A0A1S3JPC1 A0A2C9LSD6 A0A2C9LS66 U5EQB4 A0A093EZ24 A0A0B6ZN35 A0A0P7WNE8 A0A1W5B703 A0A1W5B0V3 U3I4M7 A0A3S1I1Y8 A0A093CJD4 G1NE50 A0A091HK56 A0A1J1I6J6 A0A1W5B5V9 A0A093R021 G1NE62 K7FZI2 A0A3Q3BTR5 A0A2S2Q6R3 A0A0K8TNT5 A0A3B4FB82 W5K8E1 F1NBY5 A0A091QMM2 V3ZY25 A0A099ZQS3 A0A2I0UP05 A0A093HHW2 A0A151PFQ9 A0A091MND6 A0A091U4M8 A0A1V4K7F8 A0A087QQ98 B4JBF1
A0A212FIC0 A0A1Y1KQT1 A0A2J7RC25 A0A151WQT4 E9IQK4 A0A151J196 A0A158NYX6 A0A195BVF4 A0A195FT32 A0A154PAU5 D2A2J8 A0A1W4X1R8 A0A026WDF5 A0A3L8DWS5 A0A067RD98 A0A2J7RC26 A0A088AMZ8 A0A2A3EE34 V5GV59 A0A0L7RDW4 A0A1W4WS45 A0A195D0S0 A0A1B6CAJ1 A0A0M8ZYD8 A0A1B6G5P0 A0A1B6E0S5 A0A1B6MVH7 A0A1B6L3E5 K7J9T2 A0A232EVQ4 A0A2J7RC34 A0A0J7P142 F4W5H6 E2C0H0 A0A1B6GMV3 A0A224X940 A0A069DVG0 A0A0P4VKX6 A0A0V0G4Y8 A0A023F1U4 T1HPL0 A0A1B0DJW6 A0A1L8DLW6 A0A210Q8D9 A0A034WN52 A0A1S3JNQ6 K1PL63 A0A1A9VDR9 A0A0K8TWX7 A0A1S3JPC1 A0A2C9LSD6 A0A2C9LS66 U5EQB4 A0A093EZ24 A0A0B6ZN35 A0A0P7WNE8 A0A1W5B703 A0A1W5B0V3 U3I4M7 A0A3S1I1Y8 A0A093CJD4 G1NE50 A0A091HK56 A0A1J1I6J6 A0A1W5B5V9 A0A093R021 G1NE62 K7FZI2 A0A3Q3BTR5 A0A2S2Q6R3 A0A0K8TNT5 A0A3B4FB82 W5K8E1 F1NBY5 A0A091QMM2 V3ZY25 A0A099ZQS3 A0A2I0UP05 A0A093HHW2 A0A151PFQ9 A0A091MND6 A0A091U4M8 A0A1V4K7F8 A0A087QQ98 B4JBF1
Pubmed
EMBL
BABH01005279
KZ150015
PZC74991.1
ODYU01006707
SOQ48799.1
GDQN01006292
+ More
JAT84762.1 GDQN01010013 JAT81041.1 KQ459603 KPI92946.1 AGBW02008401 OWR53476.1 GEZM01078312 JAV62771.1 NEVH01005887 PNF38393.1 KQ982821 KYQ50168.1 GL764834 EFZ17149.1 KQ980571 KYN15547.1 ADTU01004382 KQ976407 KYM91448.1 KQ981276 KYN43613.1 KQ434864 KZC09025.1 KQ971338 EFA02210.1 KK111572 KK107295 EZA46353.1 EZA53079.1 QOIP01000003 RLU24920.1 KK852532 KDR21856.1 PNF38394.1 KZ288269 PBC30015.1 GALX01004348 JAB64118.1 KQ414613 KOC69009.1 KQ977004 KYN06513.1 GEDC01030009 GEDC01026817 JAS07289.1 JAS10481.1 KQ435824 KOX72123.1 GECZ01012020 JAS57749.1 GEDC01005787 JAS31511.1 GEBQ01000042 JAT39935.1 GEBQ01021802 GEBQ01008286 JAT18175.1 JAT31691.1 AAZX01005570 NNAY01001967 OXU22427.1 PNF38392.1 LBMM01000433 KMQ98370.1 GL887645 EGI70626.1 GL451783 EFN78563.1 GECZ01006004 JAS63765.1 GFTR01007570 JAW08856.1 GBGD01000806 JAC88083.1 GDKW01001575 JAI55020.1 GECL01003025 JAP03099.1 GBBI01003731 JAC14981.1 ACPB03008272 AJVK01001177 AJVK01001178 GFDF01006719 JAV07365.1 NEDP02004628 OWF45003.1 GAKP01001941 JAC57011.1 JH817881 EKC19529.1 GDHF01033546 GDHF01009215 JAI18768.1 JAI43099.1 GANO01004328 JAB55543.1 KK378508 KFV47134.1 HACG01022937 CEK69802.1 JARO02008523 KPP62614.1 ADON01104804 RQTK01000028 RUS90612.1 KL241933 KFV12457.1 KL515096 KFO87733.1 CVRI01000038 CRK94033.1 KL432376 KFW91990.1 AGCU01075294 AGCU01075295 AGCU01075296 AGCU01075297 GGMS01004230 MBY73433.1 GDAI01002033 JAI15570.1 AADN05000096 KK664968 KFQ10699.1 KB203251 ESO85851.1 KL897682 KGL84754.1 KZ505669 PKU47785.1 KL206071 KFV78597.1 AKHW03000416 KYO47774.1 KK831060 KFP76577.1 KK416328 KFQ85381.1 LSYS01004200 OPJ80368.1 KL225807 KFM03402.1 CH916368 EDW03974.1
JAT84762.1 GDQN01010013 JAT81041.1 KQ459603 KPI92946.1 AGBW02008401 OWR53476.1 GEZM01078312 JAV62771.1 NEVH01005887 PNF38393.1 KQ982821 KYQ50168.1 GL764834 EFZ17149.1 KQ980571 KYN15547.1 ADTU01004382 KQ976407 KYM91448.1 KQ981276 KYN43613.1 KQ434864 KZC09025.1 KQ971338 EFA02210.1 KK111572 KK107295 EZA46353.1 EZA53079.1 QOIP01000003 RLU24920.1 KK852532 KDR21856.1 PNF38394.1 KZ288269 PBC30015.1 GALX01004348 JAB64118.1 KQ414613 KOC69009.1 KQ977004 KYN06513.1 GEDC01030009 GEDC01026817 JAS07289.1 JAS10481.1 KQ435824 KOX72123.1 GECZ01012020 JAS57749.1 GEDC01005787 JAS31511.1 GEBQ01000042 JAT39935.1 GEBQ01021802 GEBQ01008286 JAT18175.1 JAT31691.1 AAZX01005570 NNAY01001967 OXU22427.1 PNF38392.1 LBMM01000433 KMQ98370.1 GL887645 EGI70626.1 GL451783 EFN78563.1 GECZ01006004 JAS63765.1 GFTR01007570 JAW08856.1 GBGD01000806 JAC88083.1 GDKW01001575 JAI55020.1 GECL01003025 JAP03099.1 GBBI01003731 JAC14981.1 ACPB03008272 AJVK01001177 AJVK01001178 GFDF01006719 JAV07365.1 NEDP02004628 OWF45003.1 GAKP01001941 JAC57011.1 JH817881 EKC19529.1 GDHF01033546 GDHF01009215 JAI18768.1 JAI43099.1 GANO01004328 JAB55543.1 KK378508 KFV47134.1 HACG01022937 CEK69802.1 JARO02008523 KPP62614.1 ADON01104804 RQTK01000028 RUS90612.1 KL241933 KFV12457.1 KL515096 KFO87733.1 CVRI01000038 CRK94033.1 KL432376 KFW91990.1 AGCU01075294 AGCU01075295 AGCU01075296 AGCU01075297 GGMS01004230 MBY73433.1 GDAI01002033 JAI15570.1 AADN05000096 KK664968 KFQ10699.1 KB203251 ESO85851.1 KL897682 KGL84754.1 KZ505669 PKU47785.1 KL206071 KFV78597.1 AKHW03000416 KYO47774.1 KK831060 KFP76577.1 KK416328 KFQ85381.1 LSYS01004200 OPJ80368.1 KL225807 KFM03402.1 CH916368 EDW03974.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000235965
UP000075809
UP000078492
+ More
UP000005205 UP000078540 UP000078541 UP000076502 UP000007266 UP000192223 UP000053097 UP000279307 UP000027135 UP000005203 UP000242457 UP000053825 UP000078542 UP000053105 UP000002358 UP000215335 UP000036403 UP000007755 UP000008237 UP000015103 UP000092462 UP000242188 UP000085678 UP000005408 UP000078200 UP000076420 UP000034805 UP000192224 UP000016666 UP000271974 UP000001645 UP000183832 UP000007267 UP000264840 UP000261460 UP000018467 UP000000539 UP000030746 UP000053641 UP000053584 UP000050525 UP000190648 UP000053286 UP000001070
UP000005205 UP000078540 UP000078541 UP000076502 UP000007266 UP000192223 UP000053097 UP000279307 UP000027135 UP000005203 UP000242457 UP000053825 UP000078542 UP000053105 UP000002358 UP000215335 UP000036403 UP000007755 UP000008237 UP000015103 UP000092462 UP000242188 UP000085678 UP000005408 UP000078200 UP000076420 UP000034805 UP000192224 UP000016666 UP000271974 UP000001645 UP000183832 UP000007267 UP000264840 UP000261460 UP000018467 UP000000539 UP000030746 UP000053641 UP000053584 UP000050525 UP000190648 UP000053286 UP000001070
PRIDE
Pfam
Interpro
IPR035969
Rab-GTPase_TBC_sf
+ More
IPR021935 SGSM1/2_RBD
IPR000195 Rab-GTPase-TBC_dom
IPR034754 GEMIN8
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR011042 6-blade_b-propeller_TolB-like
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR006605 G2_nidogen/fibulin_G2F
IPR003886 NIDO_dom
IPR009017 GFP
IPR009030 Growth_fac_rcpt_cys_sf
IPR024731 EGF_dom
IPR001881 EGF-like_Ca-bd_dom
IPR000033 LDLR_classB_rpt
IPR021935 SGSM1/2_RBD
IPR000195 Rab-GTPase-TBC_dom
IPR034754 GEMIN8
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR011042 6-blade_b-propeller_TolB-like
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR006605 G2_nidogen/fibulin_G2F
IPR003886 NIDO_dom
IPR009017 GFP
IPR009030 Growth_fac_rcpt_cys_sf
IPR024731 EGF_dom
IPR001881 EGF-like_Ca-bd_dom
IPR000033 LDLR_classB_rpt
Gene 3D
ProteinModelPortal
H9JLP7
A0A2W1BL30
A0A2H1W6T5
A0A1E1WCN4
A0A1E1W2B7
A0A194PHQ3
+ More
A0A212FIC0 A0A1Y1KQT1 A0A2J7RC25 A0A151WQT4 E9IQK4 A0A151J196 A0A158NYX6 A0A195BVF4 A0A195FT32 A0A154PAU5 D2A2J8 A0A1W4X1R8 A0A026WDF5 A0A3L8DWS5 A0A067RD98 A0A2J7RC26 A0A088AMZ8 A0A2A3EE34 V5GV59 A0A0L7RDW4 A0A1W4WS45 A0A195D0S0 A0A1B6CAJ1 A0A0M8ZYD8 A0A1B6G5P0 A0A1B6E0S5 A0A1B6MVH7 A0A1B6L3E5 K7J9T2 A0A232EVQ4 A0A2J7RC34 A0A0J7P142 F4W5H6 E2C0H0 A0A1B6GMV3 A0A224X940 A0A069DVG0 A0A0P4VKX6 A0A0V0G4Y8 A0A023F1U4 T1HPL0 A0A1B0DJW6 A0A1L8DLW6 A0A210Q8D9 A0A034WN52 A0A1S3JNQ6 K1PL63 A0A1A9VDR9 A0A0K8TWX7 A0A1S3JPC1 A0A2C9LSD6 A0A2C9LS66 U5EQB4 A0A093EZ24 A0A0B6ZN35 A0A0P7WNE8 A0A1W5B703 A0A1W5B0V3 U3I4M7 A0A3S1I1Y8 A0A093CJD4 G1NE50 A0A091HK56 A0A1J1I6J6 A0A1W5B5V9 A0A093R021 G1NE62 K7FZI2 A0A3Q3BTR5 A0A2S2Q6R3 A0A0K8TNT5 A0A3B4FB82 W5K8E1 F1NBY5 A0A091QMM2 V3ZY25 A0A099ZQS3 A0A2I0UP05 A0A093HHW2 A0A151PFQ9 A0A091MND6 A0A091U4M8 A0A1V4K7F8 A0A087QQ98 B4JBF1
A0A212FIC0 A0A1Y1KQT1 A0A2J7RC25 A0A151WQT4 E9IQK4 A0A151J196 A0A158NYX6 A0A195BVF4 A0A195FT32 A0A154PAU5 D2A2J8 A0A1W4X1R8 A0A026WDF5 A0A3L8DWS5 A0A067RD98 A0A2J7RC26 A0A088AMZ8 A0A2A3EE34 V5GV59 A0A0L7RDW4 A0A1W4WS45 A0A195D0S0 A0A1B6CAJ1 A0A0M8ZYD8 A0A1B6G5P0 A0A1B6E0S5 A0A1B6MVH7 A0A1B6L3E5 K7J9T2 A0A232EVQ4 A0A2J7RC34 A0A0J7P142 F4W5H6 E2C0H0 A0A1B6GMV3 A0A224X940 A0A069DVG0 A0A0P4VKX6 A0A0V0G4Y8 A0A023F1U4 T1HPL0 A0A1B0DJW6 A0A1L8DLW6 A0A210Q8D9 A0A034WN52 A0A1S3JNQ6 K1PL63 A0A1A9VDR9 A0A0K8TWX7 A0A1S3JPC1 A0A2C9LSD6 A0A2C9LS66 U5EQB4 A0A093EZ24 A0A0B6ZN35 A0A0P7WNE8 A0A1W5B703 A0A1W5B0V3 U3I4M7 A0A3S1I1Y8 A0A093CJD4 G1NE50 A0A091HK56 A0A1J1I6J6 A0A1W5B5V9 A0A093R021 G1NE62 K7FZI2 A0A3Q3BTR5 A0A2S2Q6R3 A0A0K8TNT5 A0A3B4FB82 W5K8E1 F1NBY5 A0A091QMM2 V3ZY25 A0A099ZQS3 A0A2I0UP05 A0A093HHW2 A0A151PFQ9 A0A091MND6 A0A091U4M8 A0A1V4K7F8 A0A087QQ98 B4JBF1
PDB
5TUB
E-value=1.36024e-107,
Score=998
Ontologies
GO
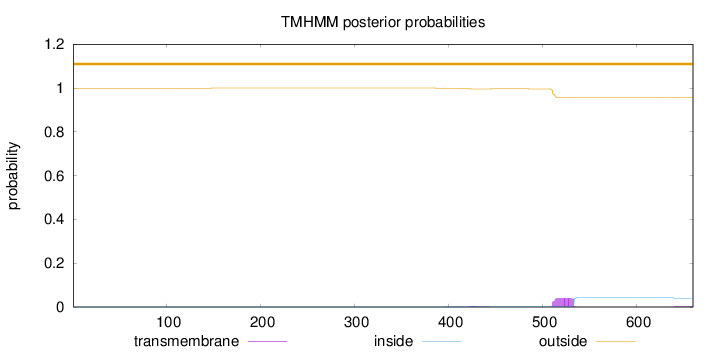
Topology
Length:
660
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.05321
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00040
outside
1 - 660
Population Genetic Test Statistics
Pi
166.238238
Theta
135.747217
Tajima's D
0.351294
CLR
0.00829
CSRT
0.472626368681566
Interpretation
Uncertain
