Gene
KWMTBOMO07046
Pre Gene Modal
BGIBMGA010451
Annotation
PREDICTED:_protein_dachsous-like_[Bombyx_mori]
Full name
Protein dachsous
Alternative Name
Adherin
Location in the cell
Nuclear Reliability : 1.667
Sequence
CDS
ATGTGGCGCGTCGTGTTGCTGGCGATGGTCGCGGCGGCGGCAAGCGAGCACGTGCGCGAGTTCGCCGTGCGAGAGAACGCACGTGCCGACACCCTCGTGGGCCACCTCGGCGACGAGCTGCCCGACGCGGCGCCGCCCTACACCATCGTCCCCGTACCCGGCAGCGCCGTCAACGACGACTTACGCGTAGATCCCCATACGGGCGAAATTCGCACCCGAGCCCCCCTTGACCGCGAGCACCGGGACCATTACGCGCTCGTCGCCATCCCCGCCTCTGGAGACAACATCAGGGTTGTCGTCCGCGTCCTCGACGAGAACGACAACGCTCCCGTCTTCCCTGCAGCCGTAATGAACGTCGAGTTCTCAGAGAACACTCCGAGGGACGTGAAAAGGAAACTTAATCCGGCCAGGGATCAAGATTTAGGGACTTTCAACACGCAACGGTACAATATAGTGTCTGGAAATACAGATAACGCGTTTAGGCTGTCCTCGCACAGGGAACGAGACGGTGTGCTGTATTTAGATTTGCAAATCAACGGATTTCTCGATAGGGAAACGACCGATCATTATCAGTTAGTTATAGAGGCGTTGGACGGCGGCACCCCACCGCTGCGGGGCACAATGACAGTGAACATTACGATATTGGACGTGAACGATAATCCCCCGGTGTTCGCTGAGAGTGTTTACTCGGCTACCATACCGGAAAACGCGACAGTCGGGACCGCGGTGCTTAAAGTGTCCGCCACCGATTCTGACGCAGGCGATAACGGTCTCATCGAGTACTCAATAAACAGGCGACAGAGTGATAAAGAGAACAATTTTAAAATCGATCCGGAAACAGGCGAGATCACTGTAAACAAGGCACTGGATTTTGAGACCAAAGATCTGCACGAATTGGTGGTGGTCGCCAGAGACAAAGGCCTCCAGCCATTAGAGACCACCGCGTTTGTTTCCATCAGAGTGACGGACGTGAACGATAATCAGCCTACTATAGACGTGATATTCTTAAGTGACGACGCCACTCCGAAAATATCAGAGTCCGCCCAATTAGATGAGTTCGTCGCGAGGATATCCGTCCACGATCCCGACTCTAAGACAGAATATTCGAACGTGAATGTGACATTGAGTGGAGGTGACGGTCACTTTGATTTGCGTACGCACGATAACATAATTTATTTGGTCGTGGTCGCGCTACCACTGGATCGCGAATCTCGATCCTCCTACACACTGAACGTTGTCGCGACGGACAAAGGTTCGCCTCCGCTGCACGCGTCTCGGATAATCTCGCTTCGAGTGACGGATGTGAACGATAATGCGCCCGCTTTTGTTGAGTCCGAATACAAAGCGAGCGTCCCGGAAGCCACCGCCCCGGGTACGCCGGTGGTTCAAGTGTCTGCTTCTGATGCCGACGAAGGCGAAAACTCGGAGATTAGGTACAGTTTGTTGACGACGCCCCAATCGGATTGGTTTTCGATTGACGAGCGCTCAGGGTTGGTCACGACTCGGTCGCGAGTCGATTGCGAGACGAACCCGATGCCTCGTTTGACGGTGGTGGCGAGGGACCGCGGCAACCCTCCGCTGTCTTCGACCGCAACGCTGCTCATCACCGTCCTGGATGTGAACGACAATGAGCCGATCTTCGACCAGTCATTCTACAACGTCACCGTGCCGGAGAGCGAAGCTGTGGGCAGCTGCATATTAAAGTGCGTCCCCAGCGCCGCTCTGGCCCCAGGCCTCGAGTGTTACGGCTACGCTATGTGCGAACTATGGGTCGGTATTTAA
Protein
MWRVVLLAMVAAAASEHVREFAVRENARADTLVGHLGDELPDAAPPYTIVPVPGSAVNDDLRVDPHTGEIRTRAPLDREHRDHYALVAIPASGDNIRVVVRVLDENDNAPVFPAAVMNVEFSENTPRDVKRKLNPARDQDLGTFNTQRYNIVSGNTDNAFRLSSHRERDGVLYLDLQINGFLDRETTDHYQLVIEALDGGTPPLRGTMTVNITILDVNDNPPVFAESVYSATIPENATVGTAVLKVSATDSDAGDNGLIEYSINRRQSDKENNFKIDPETGEITVNKALDFETKDLHELVVVARDKGLQPLETTAFVSIRVTDVNDNQPTIDVIFLSDDATPKISESAQLDEFVARISVHDPDSKTEYSNVNVTLSGGDGHFDLRTHDNIIYLVVVALPLDRESRSSYTLNVVATDKGSPPLHASRIISLRVTDVNDNAPAFVESEYKASVPEATAPGTPVVQVSASDADEGENSEIRYSLLTTPQSDWFSIDERSGLVTTRSRVDCETNPMPRLTVVARDRGNPPLSSTATLLITVLDVNDNEPIFDQSFYNVTVPESEAVGSCILKCVPSAALAPGLECYGYAMCELWVGI
Summary
Description
Required for normal morphogenesis of adult structures derived from imaginal disks (PubMed:7601355, PubMed:26073018). Plays a role in planar cell polarity and in determining body left-right asymmetry. Expression in segment H1 of the imaginal ring and interaction with Myo31DF are required to induce changes of cell shape and orientation in segment H2, which then gives rise to normal, dextral looping of the adult hindgut (PubMed:26073018).
Cadherins are calcium-dependent cell adhesion proteins.
Cadherins are calcium-dependent cell adhesion proteins.
Subunit
Interacts (via cytoplasmic region) with Myo31DF.
Miscellaneous
Overexpression in segment H1 of the imaginal ring causes aberrant gut looping in about 40% of the cases, but no phenotype is observed when both Myo31DF and ds are overexpressed.
Keywords
Calcium
Cell adhesion
Cell junction
Cell membrane
Complete proteome
Developmental protein
Glycoprotein
Membrane
Phosphoprotein
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Protein dachsous
Uniprot
H9JLP9
A0A2W1BTD6
A0A2H1WX99
A0A194QMY6
A0A194PJP2
A0A212FNF8
+ More
A0A0L7LQH3 A0A139WJW8 A0A2P8ZEV4 A0A1B0CAS0 Q17NI9 A0A067RQ34 A0A1B0DJW5 W8C4R4 A0A084VNY9 B0W508 W5J7E0 A0A2J7RC54 A0A182F4H9 A0A1I8PHT0 A0A182PME1 A0A182W9R3 A0A182STV1 A0A182RBN9 A0A182MJQ2 A0A1I8NCF2 A0A182TVC9 A0A182HVA0 A0A182WTA6 A0A1I8NCF1 A0A182Q273 A0A0K8V577 A0A182V120 A0A182YGW5 A0A0L0C915 A0A0K8TXM6 A0A182JT87 A0A2R7WTQ1 A0A0K8UZA4 A0A1J1I115 A0A1Y1JUP0 A0A182IKY5 A0A1W4WLJ5 T1IGF3 A0A1A9WW14 B4Q669 A0A3B0K446 B4P2W3 A0A0J9QT98 X2J8E9 Q24292 A0A0S1RUS1 A0A1B0GD91 B3N7T5 A0A1A9V4W2 B5DIW2 A0A1A9ZVA6 B4GPR0 A0A1W4V2Q6 B4LTP4 A0A1B0BHQ9 B4KF91 B4JDS3 B3MLQ6 A0A336LSR0 B4N0S5 A0A0M5IVU2 A0A2S2P2R1 J9LPE0 A0A2H8TRA6 A0A2P6LCN6 E9FRI4 A0A0P5QQV1 A0A0P6A4A4 A0A0P5RW78 A0A0P6FJE9 A0A0P5IJQ0 A0A0P5IPN6 A0A0P5HCT2 A0A0P6GTD1 A0A0P5QQ49 A0A0P5NCC0 A0A0P5ERX1 A0A0P5A0G7 A0A0P5AT68 A0A0P5A6D9 A0A0P5BJS1 A0A0P6AL29 A0A0P5DZ38 A0A0N7ZML0 A0A0P5DNW1 A0A0P5V9G7 A0A0P4Y9Z2 A0A164YHC3 A0A0P5BGP6 A0A0P5YF89 A0A0P5DCH6 A0A0P5P0B0 A0A088AN25 A0A2A3EJI6
A0A0L7LQH3 A0A139WJW8 A0A2P8ZEV4 A0A1B0CAS0 Q17NI9 A0A067RQ34 A0A1B0DJW5 W8C4R4 A0A084VNY9 B0W508 W5J7E0 A0A2J7RC54 A0A182F4H9 A0A1I8PHT0 A0A182PME1 A0A182W9R3 A0A182STV1 A0A182RBN9 A0A182MJQ2 A0A1I8NCF2 A0A182TVC9 A0A182HVA0 A0A182WTA6 A0A1I8NCF1 A0A182Q273 A0A0K8V577 A0A182V120 A0A182YGW5 A0A0L0C915 A0A0K8TXM6 A0A182JT87 A0A2R7WTQ1 A0A0K8UZA4 A0A1J1I115 A0A1Y1JUP0 A0A182IKY5 A0A1W4WLJ5 T1IGF3 A0A1A9WW14 B4Q669 A0A3B0K446 B4P2W3 A0A0J9QT98 X2J8E9 Q24292 A0A0S1RUS1 A0A1B0GD91 B3N7T5 A0A1A9V4W2 B5DIW2 A0A1A9ZVA6 B4GPR0 A0A1W4V2Q6 B4LTP4 A0A1B0BHQ9 B4KF91 B4JDS3 B3MLQ6 A0A336LSR0 B4N0S5 A0A0M5IVU2 A0A2S2P2R1 J9LPE0 A0A2H8TRA6 A0A2P6LCN6 E9FRI4 A0A0P5QQV1 A0A0P6A4A4 A0A0P5RW78 A0A0P6FJE9 A0A0P5IJQ0 A0A0P5IPN6 A0A0P5HCT2 A0A0P6GTD1 A0A0P5QQ49 A0A0P5NCC0 A0A0P5ERX1 A0A0P5A0G7 A0A0P5AT68 A0A0P5A6D9 A0A0P5BJS1 A0A0P6AL29 A0A0P5DZ38 A0A0N7ZML0 A0A0P5DNW1 A0A0P5V9G7 A0A0P4Y9Z2 A0A164YHC3 A0A0P5BGP6 A0A0P5YF89 A0A0P5DCH6 A0A0P5P0B0 A0A088AN25 A0A2A3EJI6
Pubmed
19121390
28756777
26354079
22118469
26227816
18362917
+ More
19820115 29403074 17510324 24845553 24495485 24438588 20920257 23761445 25315136 25244985 26108605 28004739 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7601355 18327897 18635802 26073018 26497083 15632085 23185243 18057021 21292972
19820115 29403074 17510324 24845553 24495485 24438588 20920257 23761445 25315136 25244985 26108605 28004739 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7601355 18327897 18635802 26073018 26497083 15632085 23185243 18057021 21292972
EMBL
BABH01005277
KZ150015
PZC74993.1
ODYU01011694
SOQ57596.1
KQ461194
+ More
KPJ06888.1 KQ459603 KPI92949.1 AGBW02005533 OWR55281.1 JTDY01000335 KOB77649.1 KQ971338 KYB28095.1 PYGN01000077 PSN55018.1 AJWK01004216 CH477199 EAT48227.1 KK852532 KDR21854.1 AJVK01001169 GAMC01002212 JAC04344.1 ATLV01014963 KE524999 KFB39683.1 DS231840 EDS34574.1 ADMH02002093 ETN59313.1 NEVH01005887 PNF38395.1 AXCM01004691 APCN01002503 AXCN02000870 GDHF01018416 JAI33898.1 JRES01000753 KNC28737.1 GDHF01033273 JAI19041.1 KK855479 PTY22751.1 GDHF01020649 JAI31665.1 CVRI01000038 CRK94039.1 GEZM01100129 JAV53004.1 ACPB03011283 ACPB03011284 ACPB03011285 ACPB03011286 ACPB03011287 CM000361 EDX03253.1 OUUW01000004 SPP79741.1 CM000157 EDW87170.1 CM002910 KMY87352.1 AE014134 AHN54066.1 L08811 AAF51468.3 KT935293 ALL98470.1 CCAG010015659 CH954177 EDV57261.1 CH379060 EDY70255.1 KRT04402.1 CH479187 EDW39582.1 CH940649 EDW65017.2 KRF81978.1 JXJN01014537 CH933807 EDW13074.2 CH916368 EDW03443.1 CH902620 EDV30777.1 UFQS01000093 UFQT01000093 SSW99239.1 SSX19619.1 CH963920 EDW77688.2 CP012523 ALC37924.1 GGMR01011162 MBY23781.1 ABLF02030011 ABLF02030014 GFXV01004929 MBW16734.1 MWRG01000368 PRD36340.1 GL732523 EFX90172.1 GDIQ01110813 JAL40913.1 GDIP01035182 JAM68533.1 GDIQ01095416 JAL56310.1 GDIQ01056483 JAN38254.1 GDIQ01212529 JAK39196.1 GDIQ01212528 JAK39197.1 GDIQ01229421 JAK22304.1 GDIQ01265465 GDIQ01029947 JAN64790.1 GDIQ01120959 JAL30767.1 GDIQ01154897 JAK96828.1 GDIQ01265362 JAJ86362.1 GDIP01205714 GDIP01205713 GDIP01199758 GDIP01039279 JAJ17688.1 GDIP01198310 GDIP01198309 GDIP01198308 JAJ25093.1 GDIP01203558 GDIP01203557 JAJ19844.1 GDIP01183558 JAJ39844.1 GDIP01183560 GDIP01183559 GDIP01028092 JAM75623.1 GDIP01153613 JAJ69789.1 GDIP01230657 JAI92744.1 GDIP01154663 JAJ68739.1 GDIP01102817 JAM00898.1 GDIP01230658 JAI92743.1 LRGB01000915 KZS15253.1 GDIP01184908 JAJ38494.1 GDIP01058528 GDIP01058477 JAM45187.1 GDIP01159564 JAJ63838.1 GDIQ01145352 JAL06374.1 KZ288245 PBC31191.1
KPJ06888.1 KQ459603 KPI92949.1 AGBW02005533 OWR55281.1 JTDY01000335 KOB77649.1 KQ971338 KYB28095.1 PYGN01000077 PSN55018.1 AJWK01004216 CH477199 EAT48227.1 KK852532 KDR21854.1 AJVK01001169 GAMC01002212 JAC04344.1 ATLV01014963 KE524999 KFB39683.1 DS231840 EDS34574.1 ADMH02002093 ETN59313.1 NEVH01005887 PNF38395.1 AXCM01004691 APCN01002503 AXCN02000870 GDHF01018416 JAI33898.1 JRES01000753 KNC28737.1 GDHF01033273 JAI19041.1 KK855479 PTY22751.1 GDHF01020649 JAI31665.1 CVRI01000038 CRK94039.1 GEZM01100129 JAV53004.1 ACPB03011283 ACPB03011284 ACPB03011285 ACPB03011286 ACPB03011287 CM000361 EDX03253.1 OUUW01000004 SPP79741.1 CM000157 EDW87170.1 CM002910 KMY87352.1 AE014134 AHN54066.1 L08811 AAF51468.3 KT935293 ALL98470.1 CCAG010015659 CH954177 EDV57261.1 CH379060 EDY70255.1 KRT04402.1 CH479187 EDW39582.1 CH940649 EDW65017.2 KRF81978.1 JXJN01014537 CH933807 EDW13074.2 CH916368 EDW03443.1 CH902620 EDV30777.1 UFQS01000093 UFQT01000093 SSW99239.1 SSX19619.1 CH963920 EDW77688.2 CP012523 ALC37924.1 GGMR01011162 MBY23781.1 ABLF02030011 ABLF02030014 GFXV01004929 MBW16734.1 MWRG01000368 PRD36340.1 GL732523 EFX90172.1 GDIQ01110813 JAL40913.1 GDIP01035182 JAM68533.1 GDIQ01095416 JAL56310.1 GDIQ01056483 JAN38254.1 GDIQ01212529 JAK39196.1 GDIQ01212528 JAK39197.1 GDIQ01229421 JAK22304.1 GDIQ01265465 GDIQ01029947 JAN64790.1 GDIQ01120959 JAL30767.1 GDIQ01154897 JAK96828.1 GDIQ01265362 JAJ86362.1 GDIP01205714 GDIP01205713 GDIP01199758 GDIP01039279 JAJ17688.1 GDIP01198310 GDIP01198309 GDIP01198308 JAJ25093.1 GDIP01203558 GDIP01203557 JAJ19844.1 GDIP01183558 JAJ39844.1 GDIP01183560 GDIP01183559 GDIP01028092 JAM75623.1 GDIP01153613 JAJ69789.1 GDIP01230657 JAI92744.1 GDIP01154663 JAJ68739.1 GDIP01102817 JAM00898.1 GDIP01230658 JAI92743.1 LRGB01000915 KZS15253.1 GDIP01184908 JAJ38494.1 GDIP01058528 GDIP01058477 JAM45187.1 GDIP01159564 JAJ63838.1 GDIQ01145352 JAL06374.1 KZ288245 PBC31191.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000007266
+ More
UP000245037 UP000092461 UP000008820 UP000027135 UP000092462 UP000030765 UP000002320 UP000000673 UP000235965 UP000069272 UP000095300 UP000075885 UP000075920 UP000075901 UP000075900 UP000075883 UP000095301 UP000075902 UP000075840 UP000076407 UP000075886 UP000075903 UP000076408 UP000037069 UP000075881 UP000183832 UP000075880 UP000192223 UP000015103 UP000091820 UP000000304 UP000268350 UP000002282 UP000000803 UP000092444 UP000008711 UP000078200 UP000001819 UP000092445 UP000008744 UP000192221 UP000008792 UP000092460 UP000009192 UP000001070 UP000007801 UP000007798 UP000092553 UP000007819 UP000000305 UP000076858 UP000005203 UP000242457
UP000245037 UP000092461 UP000008820 UP000027135 UP000092462 UP000030765 UP000002320 UP000000673 UP000235965 UP000069272 UP000095300 UP000075885 UP000075920 UP000075901 UP000075900 UP000075883 UP000095301 UP000075902 UP000075840 UP000076407 UP000075886 UP000075903 UP000076408 UP000037069 UP000075881 UP000183832 UP000075880 UP000192223 UP000015103 UP000091820 UP000000304 UP000268350 UP000002282 UP000000803 UP000092444 UP000008711 UP000078200 UP000001819 UP000092445 UP000008744 UP000192221 UP000008792 UP000092460 UP000009192 UP000001070 UP000007801 UP000007798 UP000092553 UP000007819 UP000000305 UP000076858 UP000005203 UP000242457
Interpro
SUPFAM
SSF49313
SSF49313
ProteinModelPortal
H9JLP9
A0A2W1BTD6
A0A2H1WX99
A0A194QMY6
A0A194PJP2
A0A212FNF8
+ More
A0A0L7LQH3 A0A139WJW8 A0A2P8ZEV4 A0A1B0CAS0 Q17NI9 A0A067RQ34 A0A1B0DJW5 W8C4R4 A0A084VNY9 B0W508 W5J7E0 A0A2J7RC54 A0A182F4H9 A0A1I8PHT0 A0A182PME1 A0A182W9R3 A0A182STV1 A0A182RBN9 A0A182MJQ2 A0A1I8NCF2 A0A182TVC9 A0A182HVA0 A0A182WTA6 A0A1I8NCF1 A0A182Q273 A0A0K8V577 A0A182V120 A0A182YGW5 A0A0L0C915 A0A0K8TXM6 A0A182JT87 A0A2R7WTQ1 A0A0K8UZA4 A0A1J1I115 A0A1Y1JUP0 A0A182IKY5 A0A1W4WLJ5 T1IGF3 A0A1A9WW14 B4Q669 A0A3B0K446 B4P2W3 A0A0J9QT98 X2J8E9 Q24292 A0A0S1RUS1 A0A1B0GD91 B3N7T5 A0A1A9V4W2 B5DIW2 A0A1A9ZVA6 B4GPR0 A0A1W4V2Q6 B4LTP4 A0A1B0BHQ9 B4KF91 B4JDS3 B3MLQ6 A0A336LSR0 B4N0S5 A0A0M5IVU2 A0A2S2P2R1 J9LPE0 A0A2H8TRA6 A0A2P6LCN6 E9FRI4 A0A0P5QQV1 A0A0P6A4A4 A0A0P5RW78 A0A0P6FJE9 A0A0P5IJQ0 A0A0P5IPN6 A0A0P5HCT2 A0A0P6GTD1 A0A0P5QQ49 A0A0P5NCC0 A0A0P5ERX1 A0A0P5A0G7 A0A0P5AT68 A0A0P5A6D9 A0A0P5BJS1 A0A0P6AL29 A0A0P5DZ38 A0A0N7ZML0 A0A0P5DNW1 A0A0P5V9G7 A0A0P4Y9Z2 A0A164YHC3 A0A0P5BGP6 A0A0P5YF89 A0A0P5DCH6 A0A0P5P0B0 A0A088AN25 A0A2A3EJI6
A0A0L7LQH3 A0A139WJW8 A0A2P8ZEV4 A0A1B0CAS0 Q17NI9 A0A067RQ34 A0A1B0DJW5 W8C4R4 A0A084VNY9 B0W508 W5J7E0 A0A2J7RC54 A0A182F4H9 A0A1I8PHT0 A0A182PME1 A0A182W9R3 A0A182STV1 A0A182RBN9 A0A182MJQ2 A0A1I8NCF2 A0A182TVC9 A0A182HVA0 A0A182WTA6 A0A1I8NCF1 A0A182Q273 A0A0K8V577 A0A182V120 A0A182YGW5 A0A0L0C915 A0A0K8TXM6 A0A182JT87 A0A2R7WTQ1 A0A0K8UZA4 A0A1J1I115 A0A1Y1JUP0 A0A182IKY5 A0A1W4WLJ5 T1IGF3 A0A1A9WW14 B4Q669 A0A3B0K446 B4P2W3 A0A0J9QT98 X2J8E9 Q24292 A0A0S1RUS1 A0A1B0GD91 B3N7T5 A0A1A9V4W2 B5DIW2 A0A1A9ZVA6 B4GPR0 A0A1W4V2Q6 B4LTP4 A0A1B0BHQ9 B4KF91 B4JDS3 B3MLQ6 A0A336LSR0 B4N0S5 A0A0M5IVU2 A0A2S2P2R1 J9LPE0 A0A2H8TRA6 A0A2P6LCN6 E9FRI4 A0A0P5QQV1 A0A0P6A4A4 A0A0P5RW78 A0A0P6FJE9 A0A0P5IJQ0 A0A0P5IPN6 A0A0P5HCT2 A0A0P6GTD1 A0A0P5QQ49 A0A0P5NCC0 A0A0P5ERX1 A0A0P5A0G7 A0A0P5AT68 A0A0P5A6D9 A0A0P5BJS1 A0A0P6AL29 A0A0P5DZ38 A0A0N7ZML0 A0A0P5DNW1 A0A0P5V9G7 A0A0P4Y9Z2 A0A164YHC3 A0A0P5BGP6 A0A0P5YF89 A0A0P5DCH6 A0A0P5P0B0 A0A088AN25 A0A2A3EJI6
PDB
5T9T
E-value=3.31593e-67,
Score=649
Ontologies
PATHWAY
GO
GO:0016021
GO:0007156
GO:0005886
GO:0005509
GO:0007155
GO:0005887
GO:0045296
GO:0044331
GO:0016339
GO:0045317
GO:0045198
GO:0007480
GO:0007157
GO:0001737
GO:0090176
GO:0035222
GO:0090175
GO:0035159
GO:0007164
GO:0048592
GO:0016318
GO:0001736
GO:0098609
GO:0007476
GO:0042067
GO:0030010
GO:0035331
GO:0060071
GO:0016327
GO:0000904
GO:0008283
GO:0030054
GO:0018149
GO:0090251
GO:0035332
GO:0016020
GO:0005515
GO:0004319
GO:0004320
GO:0016295
GO:0016296
Topology
Subcellular location
Cell membrane
Colocalizes with Myo32DF at cell contact sites. With evidence from 5 publications.
Cell junction Colocalizes with Myo32DF at cell contact sites. With evidence from 5 publications.
Cell junction Colocalizes with Myo32DF at cell contact sites. With evidence from 5 publications.
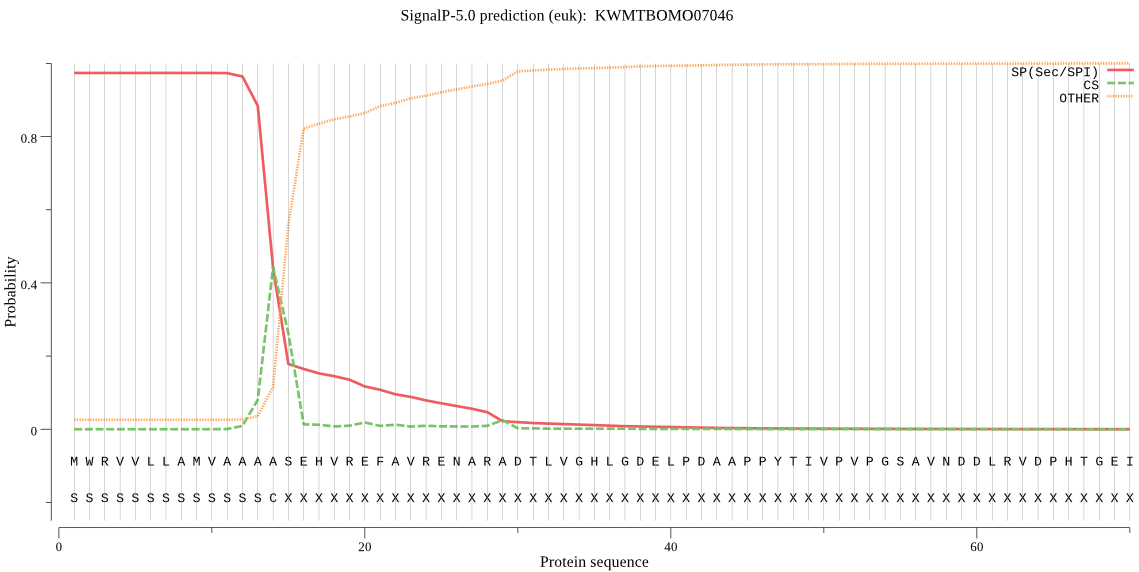
SignalP
Position: 1 - 14,
Likelihood: 0.973414
Length:
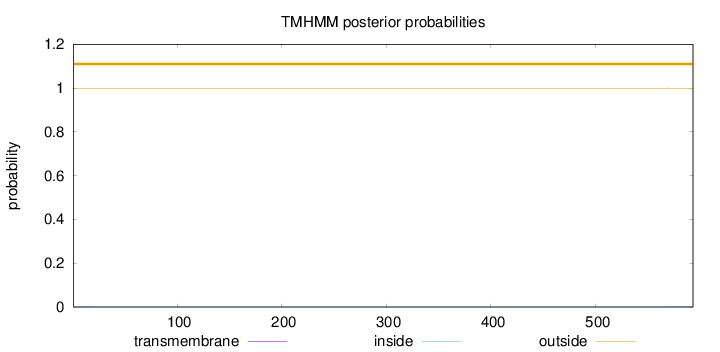
593
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01154
Exp number, first 60 AAs:
0.00252
Total prob of N-in:
0.00053
outside
1 - 593
Population Genetic Test Statistics
Pi
205.021873
Theta
175.623859
Tajima's D
0.990177
CLR
0.204251
CSRT
0.657967101644918
Interpretation
Uncertain
