Gene
KWMTBOMO07044
Pre Gene Modal
BGIBMGA010513
Annotation
PREDICTED:_glutamate_transporter_isoform_X1_[Bombyx_mori]
Full name
Amino acid transporter
Location in the cell
PlasmaMembrane Reliability : 4.916
Sequence
CDS
ATGAATCATATCATTACACTTCTAGAGTACTTGTTCGCTAAGATGGGGCCCGGTGATCGTGGGCCGAAGACTACAGAGGAACGATCAGCACCATTAGAGCCAACCGGCTGGAGGAAGTGCGTTATCCATAATACCATGCTTGTGGTCACTCTCGTCGGCGTCGTCGCTGGGATTGCATTAGGGTTCGGTTTGCGGCCCTATCAACTAGGTCCGGACACGTTGGTCCTCATCTCATATCCGGGCGAGCTCTTCATGAGGCTACTGAAACTCATGATACTGCCGCTGATCATAGCCAGCCTGATCGCCGGTTCCGCAAGTCTTAACGCTAAGATGAGCGGGAGGATCGCAGTGAGAACATTAGTTTATTTCATTTTAACTTCAATGTTCAACGCGTTTCTTGGTATCGTAATGGCGGTCCTGATCCATCCCGGACAGCCGGAACTGAAGGAGGACGTTCTGGTCGCTGCCGGCAGTAAGAAAGATCACAGCATTCTGGATAGCTTGCTGGATATAGGAAGAAACATATTCCCCGATAACATAGTGCAGGCGGCTTTCCAGCAAGCGCACACGGTGTACGTCGAGGAGACCACGTTGCTTGCCAAGAATTCAACCGAGAATGGCACTGTGCCGACTTTGGTACGAGTCGTGTCTTACAGATCTGGCACAAATACTTTAGGTCTGGTATTCTTCTGCCTGGTTTTTGGAAGTCTTCTCGGATCTTTGGGCGCAAAAGGCCAAGTAGTCATAGATTTCTTTCAAGCTATATTTGAAGTGATCATGAAAATGGTAGCTGGAGTGATGTGGTTCACCCCGTTTGGTGTCAGCAGTATTATAGCTGGGAAGATATTGGGTGTAGCAGATGTGGCACACGTGATGTCCCAGTTGGCATGGTTCATAGCGACAGTGGCCGTGGGAGTGTTCCTTTATCAGCTCATAGTGATGCAACTGATATATTTCATAGTCGTGAAAAAGAATCCGTATAAGTTCTACTGGGGACTGTCGCACGCTATGCTAACTGCCTCAGCTACGGCGTCAACTGCAGCAGCGTTGCCCGTCACCTTCAAAGCAATGGAAGGTCCACTCAGGATCGATGTCCGCATCACTAGGTTCGTTCTTCCGATTGGCTGTAACATCAACATGGACGGCACCGCTCTCTTCATAGCGATAGCCGCGGTCTTCATATGCCAAATGAACAATATGACTTTGGGTTTTGCTCAATTGGCAACTATATTTTTGACGTCGACGGCCGCGTCTCTGTCGTCGGCGTCGGTACCGTCGGCGGCGCTGGTGTTGCTGCTGGTGGTGCTGTCCTCAGTGGACGCGCCTGCGCAGGACGTGTCTCTGTTATTCGCTGTGGACTGGCTCGTTGATAGGATTCGCACAACGAACAACATGCTCGGCGACTGCTACGCGGCCGCCGTCGTGGAGCATCTGTCTAAGAACGAGCTGATGGCGTGCGATGCGGCTTCCATTGAAGCTACGACCGGGATAACTCCGAGCACAGAAGAAGCCTTAACCACGCCAGGGAAGCAGTCGATCGTATCGGACGACATCGTCATTGACATGCACAATCACAACAACGTCGAAAGGATTTAG
Protein
MNHIITLLEYLFAKMGPGDRGPKTTEERSAPLEPTGWRKCVIHNTMLVVTLVGVVAGIALGFGLRPYQLGPDTLVLISYPGELFMRLLKLMILPLIIASLIAGSASLNAKMSGRIAVRTLVYFILTSMFNAFLGIVMAVLIHPGQPELKEDVLVAAGSKKDHSILDSLLDIGRNIFPDNIVQAAFQQAHTVYVEETTLLAKNSTENGTVPTLVRVVSYRSGTNTLGLVFFCLVFGSLLGSLGAKGQVVIDFFQAIFEVIMKMVAGVMWFTPFGVSSIIAGKILGVADVAHVMSQLAWFIATVAVGVFLYQLIVMQLIYFIVVKKNPYKFYWGLSHAMLTASATASTAAALPVTFKAMEGPLRIDVRITRFVLPIGCNINMDGTALFIAIAAVFICQMNNMTLGFAQLATIFLTSTAASLSSASVPSAALVLLLVVLSSVDAPAQDVSLLFAVDWLVDRIRTTNNMLGDCYAAAVVEHLSKNELMACDAASIEATTGITPSTEEALTTPGKQSIVSDDIVIDMHNHNNVERI
Summary
Similarity
Belongs to the dicarboxylate/amino acid:cation symporter (DAACS) (TC 2.A.23) family.
Feature
chain Amino acid transporter
Uniprot
G9BIL4
A0A2A4K209
A0A2W1B7G6
A0A0G3VGU7
A0A212FNJ5
A0A194PHQ7
+ More
A0A194QTL3 A0A2H1VPD9 A0A0L7LU62 K7J9T1 Q29LM2 B4KF87 A0A1I8MMG5 A0A1A9ZW30 A0A3B0JHH2 A0A232EQM6 B4P2W5 A0A0M4EPE5 A0A0A1X3B4 E1JHQ6 A0A0R3NT53 A0A0L0C902 A0A034V3C4 A0A0K8UX96 A0A0Q9XH56 B4GPQ9 B4LTP5 A0A026WAQ6 A0A3L8DWG6 A0A154PRZ9 A0A1B0GEJ4 A0A158NYX2 A0A1I8MMG4 A0A195D2C4 A0A3B0JGB2 F4W5G1 A0A151WQM1 A0A1A9V0U6 A0A151J1J7 A0A0A1WJX6 A0A0R1DIU6 Q9U9K1 B4JDS5 A0A034V8A8 A0A1I8PTY0 Q9VPS6 A0A0Q9WB85 B3N7T7 E2C0G5 E2A616 A0A0J9QU80 A0A0K8VXC2 A0A0C9QWV7 B3MLQ8 W8ASF2 A0A0L7RDW9 A0A088AN22 B4N0S3 A0A0Q5W8K4 A0A195BTN2 A0A195FT79 A0A0P9BMM7 A0A1B0CFK4 B4ID06 B4Q6I9 A0A0N0BEN2 A0A1W4UQP8 A0A2A3EHC8 A0A0Q9WRF8 A0A0J7LB87 A0A182NE58 A0A182QGB1 A0A182YGW8 A0A182RBP3 A0A182JWX4 A0A182W9R7 A0A182PMD7 A0A182F4I2 A0A1A9WW12 B0W510 Q5TQ49 A0A1S4GWL7 A0A182V4E3 A0A1Q3FSF8 A0A182HVA5 A0A182WTB2 A0A182L272 Q17NI7 A0A1S4EWM8 A0A023ETG8 A0A023EUI7 D7ELY7 A0A1Y0AWS8 A0A182S9E1 A0A139W8W4 A0A182M8I5 T1P7I5 A0A182UL30 W5J4D2 A0A1S4GWN4
A0A194QTL3 A0A2H1VPD9 A0A0L7LU62 K7J9T1 Q29LM2 B4KF87 A0A1I8MMG5 A0A1A9ZW30 A0A3B0JHH2 A0A232EQM6 B4P2W5 A0A0M4EPE5 A0A0A1X3B4 E1JHQ6 A0A0R3NT53 A0A0L0C902 A0A034V3C4 A0A0K8UX96 A0A0Q9XH56 B4GPQ9 B4LTP5 A0A026WAQ6 A0A3L8DWG6 A0A154PRZ9 A0A1B0GEJ4 A0A158NYX2 A0A1I8MMG4 A0A195D2C4 A0A3B0JGB2 F4W5G1 A0A151WQM1 A0A1A9V0U6 A0A151J1J7 A0A0A1WJX6 A0A0R1DIU6 Q9U9K1 B4JDS5 A0A034V8A8 A0A1I8PTY0 Q9VPS6 A0A0Q9WB85 B3N7T7 E2C0G5 E2A616 A0A0J9QU80 A0A0K8VXC2 A0A0C9QWV7 B3MLQ8 W8ASF2 A0A0L7RDW9 A0A088AN22 B4N0S3 A0A0Q5W8K4 A0A195BTN2 A0A195FT79 A0A0P9BMM7 A0A1B0CFK4 B4ID06 B4Q6I9 A0A0N0BEN2 A0A1W4UQP8 A0A2A3EHC8 A0A0Q9WRF8 A0A0J7LB87 A0A182NE58 A0A182QGB1 A0A182YGW8 A0A182RBP3 A0A182JWX4 A0A182W9R7 A0A182PMD7 A0A182F4I2 A0A1A9WW12 B0W510 Q5TQ49 A0A1S4GWL7 A0A182V4E3 A0A1Q3FSF8 A0A182HVA5 A0A182WTB2 A0A182L272 Q17NI7 A0A1S4EWM8 A0A023ETG8 A0A023EUI7 D7ELY7 A0A1Y0AWS8 A0A182S9E1 A0A139W8W4 A0A182M8I5 T1P7I5 A0A182UL30 W5J4D2 A0A1S4GWN4
Pubmed
19121390
28756777
22118469
26354079
26227816
20075255
+ More
15632085 17994087 25315136 28648823 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 25348373 18057021 24508170 30249741 21347285 21719571 9989583 26109357 26109356 20798317 22936249 24495485 25244985 12364791 20966253 17510324 24945155 26483478 18362917 19820115 28341416 20920257 23761445
15632085 17994087 25315136 28648823 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 25348373 18057021 24508170 30249741 21347285 21719571 9989583 26109357 26109356 20798317 22936249 24495485 25244985 12364791 20966253 17510324 24945155 26483478 18362917 19820115 28341416 20920257 23761445
EMBL
BABH01005263
BABH01005264
BABH01005265
HQ588718
AEL88626.1
NWSH01000294
+ More
PCG77672.1 KZ150312 PZC71458.1 KP657651 AKL78876.1 AGBW02005533 OWR55279.1 KQ459603 KPI92951.1 KQ461194 KPJ06886.1 ODYU01003656 SOQ42701.1 JTDY01000080 KOB78987.1 AAZX01003632 CH379060 EAL34023.2 CH933807 EDW13070.2 OUUW01000004 SPP79742.1 NNAY01002748 OXU20632.1 CM000157 EDW87172.2 CP012523 ALC38445.1 GBXI01008946 JAD05346.1 AE014134 ACZ94136.1 KRT04401.1 JRES01000753 KNC28735.1 GAKP01021177 GAKP01021176 JAC37776.1 GDHF01033296 GDHF01027062 GDHF01021037 GDHF01018407 GDHF01011379 GDHF01010536 GDHF01007822 JAI19018.1 JAI25252.1 JAI31277.1 JAI33907.1 JAI40935.1 JAI41778.1 JAI44492.1 KRG03569.1 CH479187 EDW39581.1 CH940649 EDW65018.2 KK107295 EZA53063.1 QOIP01000003 RLU24415.1 KQ435102 KZC14671.1 CCAG010012288 ADTU01004342 KQ977004 KYN06529.1 SPP79743.1 GL887645 EGI70611.1 KQ982821 KYQ50154.1 KQ980571 KYN15532.1 GBXI01015507 JAC98784.1 KRJ97048.1 AF166000 AAD47830.1 CH916368 EDW03445.1 GAKP01021174 JAC37778.1 AAF51466.1 KRF81980.1 CH954177 EDV57263.2 GL451783 EFN78558.1 GL437108 EFN71075.1 CM002910 KMY87354.1 GDHF01008776 JAI43538.1 GBYB01000180 JAG69947.1 CH902620 EDV30779.2 GAMC01019042 GAMC01019035 JAB87513.1 KQ414613 KOC69014.1 CH963920 EDW77686.2 KQS69936.1 KQ976407 KYM91461.1 KQ981276 KYN43631.1 KPU73031.1 AJWK01010139 CH480829 EDW45432.1 CM000361 EDX03255.1 KMY87355.1 KQ435824 KOX72112.1 KZ288245 PBC31193.1 KRF98721.1 LBMM01000048 KMR05172.1 AXCN02000870 DS231840 EDS34576.1 AAAB01008964 EAL39706.3 GFDL01004663 JAV30382.1 APCN01002503 CH477199 EAT48229.1 JXUM01015453 JXUM01015454 JXUM01035309 GAPW01000958 KQ561054 KQ560430 JAC12640.1 KXJ79853.1 KXJ82443.1 GAPW01000976 JAC12622.1 KQ972953 EFA12466.2 KY921800 ART29389.1 KXZ75717.1 AXCM01000141 KA644424 AFP59053.1 ADMH02002093 ETN59317.1
PCG77672.1 KZ150312 PZC71458.1 KP657651 AKL78876.1 AGBW02005533 OWR55279.1 KQ459603 KPI92951.1 KQ461194 KPJ06886.1 ODYU01003656 SOQ42701.1 JTDY01000080 KOB78987.1 AAZX01003632 CH379060 EAL34023.2 CH933807 EDW13070.2 OUUW01000004 SPP79742.1 NNAY01002748 OXU20632.1 CM000157 EDW87172.2 CP012523 ALC38445.1 GBXI01008946 JAD05346.1 AE014134 ACZ94136.1 KRT04401.1 JRES01000753 KNC28735.1 GAKP01021177 GAKP01021176 JAC37776.1 GDHF01033296 GDHF01027062 GDHF01021037 GDHF01018407 GDHF01011379 GDHF01010536 GDHF01007822 JAI19018.1 JAI25252.1 JAI31277.1 JAI33907.1 JAI40935.1 JAI41778.1 JAI44492.1 KRG03569.1 CH479187 EDW39581.1 CH940649 EDW65018.2 KK107295 EZA53063.1 QOIP01000003 RLU24415.1 KQ435102 KZC14671.1 CCAG010012288 ADTU01004342 KQ977004 KYN06529.1 SPP79743.1 GL887645 EGI70611.1 KQ982821 KYQ50154.1 KQ980571 KYN15532.1 GBXI01015507 JAC98784.1 KRJ97048.1 AF166000 AAD47830.1 CH916368 EDW03445.1 GAKP01021174 JAC37778.1 AAF51466.1 KRF81980.1 CH954177 EDV57263.2 GL451783 EFN78558.1 GL437108 EFN71075.1 CM002910 KMY87354.1 GDHF01008776 JAI43538.1 GBYB01000180 JAG69947.1 CH902620 EDV30779.2 GAMC01019042 GAMC01019035 JAB87513.1 KQ414613 KOC69014.1 CH963920 EDW77686.2 KQS69936.1 KQ976407 KYM91461.1 KQ981276 KYN43631.1 KPU73031.1 AJWK01010139 CH480829 EDW45432.1 CM000361 EDX03255.1 KMY87355.1 KQ435824 KOX72112.1 KZ288245 PBC31193.1 KRF98721.1 LBMM01000048 KMR05172.1 AXCN02000870 DS231840 EDS34576.1 AAAB01008964 EAL39706.3 GFDL01004663 JAV30382.1 APCN01002503 CH477199 EAT48229.1 JXUM01015453 JXUM01015454 JXUM01035309 GAPW01000958 KQ561054 KQ560430 JAC12640.1 KXJ79853.1 KXJ82443.1 GAPW01000976 JAC12622.1 KQ972953 EFA12466.2 KY921800 ART29389.1 KXZ75717.1 AXCM01000141 KA644424 AFP59053.1 ADMH02002093 ETN59317.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000002358 UP000001819 UP000009192 UP000095301 UP000092445 UP000268350 UP000215335 UP000002282 UP000092553 UP000000803 UP000037069 UP000008744 UP000008792 UP000053097 UP000279307 UP000076502 UP000092444 UP000005205 UP000078542 UP000007755 UP000075809 UP000078200 UP000078492 UP000001070 UP000095300 UP000008711 UP000008237 UP000000311 UP000007801 UP000053825 UP000005203 UP000007798 UP000078540 UP000078541 UP000092461 UP000001292 UP000000304 UP000053105 UP000192221 UP000242457 UP000036403 UP000075884 UP000075886 UP000076408 UP000075900 UP000075881 UP000075920 UP000075885 UP000069272 UP000091820 UP000002320 UP000007062 UP000075903 UP000075840 UP000076407 UP000075882 UP000008820 UP000069940 UP000249989 UP000007266 UP000075901 UP000075883 UP000075902 UP000000673
UP000002358 UP000001819 UP000009192 UP000095301 UP000092445 UP000268350 UP000215335 UP000002282 UP000092553 UP000000803 UP000037069 UP000008744 UP000008792 UP000053097 UP000279307 UP000076502 UP000092444 UP000005205 UP000078542 UP000007755 UP000075809 UP000078200 UP000078492 UP000001070 UP000095300 UP000008711 UP000008237 UP000000311 UP000007801 UP000053825 UP000005203 UP000007798 UP000078540 UP000078541 UP000092461 UP000001292 UP000000304 UP000053105 UP000192221 UP000242457 UP000036403 UP000075884 UP000075886 UP000076408 UP000075900 UP000075881 UP000075920 UP000075885 UP000069272 UP000091820 UP000002320 UP000007062 UP000075903 UP000075840 UP000076407 UP000075882 UP000008820 UP000069940 UP000249989 UP000007266 UP000075901 UP000075883 UP000075902 UP000000673
Pfam
PF00375 SDF
Interpro
SUPFAM
SSF118215
SSF118215
Gene 3D
ProteinModelPortal
G9BIL4
A0A2A4K209
A0A2W1B7G6
A0A0G3VGU7
A0A212FNJ5
A0A194PHQ7
+ More
A0A194QTL3 A0A2H1VPD9 A0A0L7LU62 K7J9T1 Q29LM2 B4KF87 A0A1I8MMG5 A0A1A9ZW30 A0A3B0JHH2 A0A232EQM6 B4P2W5 A0A0M4EPE5 A0A0A1X3B4 E1JHQ6 A0A0R3NT53 A0A0L0C902 A0A034V3C4 A0A0K8UX96 A0A0Q9XH56 B4GPQ9 B4LTP5 A0A026WAQ6 A0A3L8DWG6 A0A154PRZ9 A0A1B0GEJ4 A0A158NYX2 A0A1I8MMG4 A0A195D2C4 A0A3B0JGB2 F4W5G1 A0A151WQM1 A0A1A9V0U6 A0A151J1J7 A0A0A1WJX6 A0A0R1DIU6 Q9U9K1 B4JDS5 A0A034V8A8 A0A1I8PTY0 Q9VPS6 A0A0Q9WB85 B3N7T7 E2C0G5 E2A616 A0A0J9QU80 A0A0K8VXC2 A0A0C9QWV7 B3MLQ8 W8ASF2 A0A0L7RDW9 A0A088AN22 B4N0S3 A0A0Q5W8K4 A0A195BTN2 A0A195FT79 A0A0P9BMM7 A0A1B0CFK4 B4ID06 B4Q6I9 A0A0N0BEN2 A0A1W4UQP8 A0A2A3EHC8 A0A0Q9WRF8 A0A0J7LB87 A0A182NE58 A0A182QGB1 A0A182YGW8 A0A182RBP3 A0A182JWX4 A0A182W9R7 A0A182PMD7 A0A182F4I2 A0A1A9WW12 B0W510 Q5TQ49 A0A1S4GWL7 A0A182V4E3 A0A1Q3FSF8 A0A182HVA5 A0A182WTB2 A0A182L272 Q17NI7 A0A1S4EWM8 A0A023ETG8 A0A023EUI7 D7ELY7 A0A1Y0AWS8 A0A182S9E1 A0A139W8W4 A0A182M8I5 T1P7I5 A0A182UL30 W5J4D2 A0A1S4GWN4
A0A194QTL3 A0A2H1VPD9 A0A0L7LU62 K7J9T1 Q29LM2 B4KF87 A0A1I8MMG5 A0A1A9ZW30 A0A3B0JHH2 A0A232EQM6 B4P2W5 A0A0M4EPE5 A0A0A1X3B4 E1JHQ6 A0A0R3NT53 A0A0L0C902 A0A034V3C4 A0A0K8UX96 A0A0Q9XH56 B4GPQ9 B4LTP5 A0A026WAQ6 A0A3L8DWG6 A0A154PRZ9 A0A1B0GEJ4 A0A158NYX2 A0A1I8MMG4 A0A195D2C4 A0A3B0JGB2 F4W5G1 A0A151WQM1 A0A1A9V0U6 A0A151J1J7 A0A0A1WJX6 A0A0R1DIU6 Q9U9K1 B4JDS5 A0A034V8A8 A0A1I8PTY0 Q9VPS6 A0A0Q9WB85 B3N7T7 E2C0G5 E2A616 A0A0J9QU80 A0A0K8VXC2 A0A0C9QWV7 B3MLQ8 W8ASF2 A0A0L7RDW9 A0A088AN22 B4N0S3 A0A0Q5W8K4 A0A195BTN2 A0A195FT79 A0A0P9BMM7 A0A1B0CFK4 B4ID06 B4Q6I9 A0A0N0BEN2 A0A1W4UQP8 A0A2A3EHC8 A0A0Q9WRF8 A0A0J7LB87 A0A182NE58 A0A182QGB1 A0A182YGW8 A0A182RBP3 A0A182JWX4 A0A182W9R7 A0A182PMD7 A0A182F4I2 A0A1A9WW12 B0W510 Q5TQ49 A0A1S4GWL7 A0A182V4E3 A0A1Q3FSF8 A0A182HVA5 A0A182WTB2 A0A182L272 Q17NI7 A0A1S4EWM8 A0A023ETG8 A0A023EUI7 D7ELY7 A0A1Y0AWS8 A0A182S9E1 A0A139W8W4 A0A182M8I5 T1P7I5 A0A182UL30 W5J4D2 A0A1S4GWN4
PDB
5MJU
E-value=1.84567e-89,
Score=841
Ontologies
GO
Topology
Subcellular location
Membrane
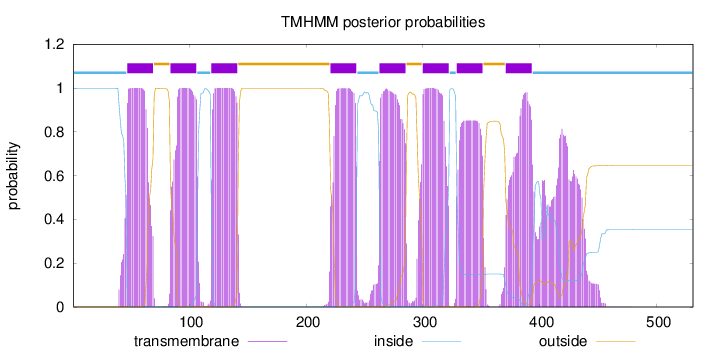
Length:
531
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
197.42117
Exp number, first 60 AAs:
15.72309
Total prob of N-in:
0.99875
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 69
outside
70 - 83
TMhelix
84 - 106
inside
107 - 118
TMhelix
119 - 141
outside
142 - 220
TMhelix
221 - 243
inside
244 - 262
TMhelix
263 - 285
outside
286 - 299
TMhelix
300 - 322
inside
323 - 328
TMhelix
329 - 351
outside
352 - 370
TMhelix
371 - 393
inside
394 - 531
Population Genetic Test Statistics
Pi
227.733444
Theta
179.039465
Tajima's D
1.444082
CLR
0.236874
CSRT
0.770611469426529
Interpretation
Uncertain
