Gene
KWMTBOMO07041
Pre Gene Modal
BGIBMGA010454
Annotation
integrin_alpha-PS3-like_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.233
Sequence
CDS
ATGTATAGATCAATTATCTATTTAAGCGTTTTACTGTGTTGTGAATTTGTATCCGGTATTGACGTATTCCACGAGGGATCAAAAATAACGTTTCACCCCAACGATGGAGGGTCAGGTTATTTTGGTTACAGCGTCTCTCTGAGCTCCTACGGTCTCGTAGTTGGTGCGCCAAAAGCTGTGGCCAAAATGCCCTCCTCCGTTTCATCAGGTTTGGTCTACAACTGTCCTCTAAAGGACACGATGGATGCTAGCAACGTTACTTGCAATGTAATTAGTCTGGGCAGCTCGCGATATCTGGGTTACGTGTCGTTTTTAGACGAAATGATCAAGGATGACATGTGGTTCGGTGCAACCATAGCCCCCATGTCGAAGAACAAACTATTGATCTGCGCTCCTCGTTGGCTGGTCACGTACAAACGCAAGCATTACTTGGCGAACGGAGTGTGTTTTTTACACAAAAACGAACGAGACTGGATATTGTATCCATTGAGGGACATGGATCGCCAGGCTTTCGTGGTAAACGGTGACAGGATTGAATACGGGGAATACGGTCTCCATGTCAACTTCTACGCTTACGGTCAAGCGGGAATGTCGACCAAAGTGACCGAAGACAATTACGTGATCGTGGGAGCCCCTGGACTACTGCAATGGACAGGCGGTATCGTGAGTTTCAAATACAGCCAGGACGAGGAGAGCTCGTATATCAGCAAACAGACAACTTCGAATCCTTACTACACCCGGGATCTTGAACCTTATAGCTATTTCGGTTACAGTGTAGAGTCTGGGATATTCGAACCAAACGGGAGCGTATTGTATGTAAGCGGAGGTCCGCGTGCACATATGGGCTATGGAGAGGTTTTAGTCTTCAAACCAGTTCAAAGAGAGAAGGATCCCTTGAATATCTTGGCGAAATTGGTAGGACCGCATCTCGGAGCTTATTTTGGAGCTAGTCTTTGCTGTACAGACATTGATGGTGACGGCCGATCAGATCTGCTGGTTGGAGCACCAACGTATGTGAAGAAAGACGGTGGACTGCCTTTTGATCAAGGCGCCGTATTCGTTTACATGACTCGAGAGAAAGATTCGAGCTTTGTTTTAGAAGAAGCGGGTATGGTCCTCGGCTCTGGTGAGAGCGGAGCTCATTTTGGGATAGCTATTGCTGATTTGGGTGATATTGATGGGGATGGCTATAAAGATATTGCCATAGGCGCACCCTGGGAGGATGAAGGAACTGGCGCTGTCTACATTTACAAAGGACATAAGAAAGGTTTAAGAAGTAATCACATTCAGAGGATATCGCCAGAAGGCGCAAGAAGCTTTGGAATGTCAATATCTAAAGGATACGACGTGGATAACAATAACTGTAGTGATCTAGCCGTGGGTGCTCATGAAAGTGCTGAAGTTTACCTCTTCAGATGCATTCCATCCATGGACGTTCACGCTCAGATCGAAGTTCCAGATGCCATCAATTTACCACAGAACACAACCGAATTCACAGCGCTTGCCTGTATTGAGATACCCGATAAGCCTTTGTGGCGACACGTTAAATTATATATAAAATCACGAATAATCGTTGACCCGGAACAAAACAGAGCGCAGGTATCGGGCGATTCGGAAACTATTGTAGTGGCGAGGCCCGGAGACAAACAATGCACTGAACGGACTATTGCGGTTAAAGTAAGTTACAGTATTGTTACACTCGATCCAGTATTCAGTGTGCTTAGTACGAGTTTTTAA
Protein
MYRSIIYLSVLLCCEFVSGIDVFHEGSKITFHPNDGGSGYFGYSVSLSSYGLVVGAPKAVAKMPSSVSSGLVYNCPLKDTMDASNVTCNVISLGSSRYLGYVSFLDEMIKDDMWFGATIAPMSKNKLLICAPRWLVTYKRKHYLANGVCFLHKNERDWILYPLRDMDRQAFVVNGDRIEYGEYGLHVNFYAYGQAGMSTKVTEDNYVIVGAPGLLQWTGGIVSFKYSQDEESSYISKQTTSNPYYTRDLEPYSYFGYSVESGIFEPNGSVLYVSGGPRAHMGYGEVLVFKPVQREKDPLNILAKLVGPHLGAYFGASLCCTDIDGDGRSDLLVGAPTYVKKDGGLPFDQGAVFVYMTREKDSSFVLEEAGMVLGSGESGAHFGIAIADLGDIDGDGYKDIAIGAPWEDEGTGAVYIYKGHKKGLRSNHIQRISPEGARSFGMSISKGYDVDNNNCSDLAVGAHESAEVYLFRCIPSMDVHAQIEVPDAINLPQNTTEFTALACIEIPDKPLWRHVKLYIKSRIIVDPEQNRAQVSGDSETIVVARPGDKQCTERTIAVKVSYSIVTLDPVFSVLSTSF
Summary
Subunit
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the integrin alpha chain family.
Uniprot
A0A076JQH6
H9JLQ2
Q1G0S5
A0A2A4JPM1
A0A194QPI3
A0A194PIS8
+ More
A0A212F4X5 A0A2W1B894 A0A2H1VUW8 Q86G86 A0A3S2NWJ7 A0A2H1V8F5 A0A0L7LUC9 A0A2J7QMA1 A0A3R7SKY0 A0A0A0PGH3 A0A087UCA8 A0A2Z5WPE6 A0A147BHT3 A0A034VIP4 A0A0K8V796 A0A0K8VM67 A0A0K8VIA7 A0A0A1XJR9 A0A0A1X2M1 Q16R90 Q16JC5 W8BH52 W8AK36 A0A182HAI7 A0A1I8N608 A0A293N3R2 A0A1Y1LAY1 A0A1W4WBX2 A0A3Q4N2Y5 A0A1Z5LDZ0 A0A3Q2VM08 F6R3Z0 A0A3B4YNS0 A0A0P4VVE3 A0A3B4U9I1 A0A1Y1LCJ3 A0A182Y859 A0A1A9VE28 A0A3Q3FQK3 A0A1Y1L8R1 I3JIG3 Q8BQ25 A0A3B4FXJ4 A0A195E483 A0A1A9WWH1 A0A151X2M7 A0A1B0FDC0 A0A1B0A219 A0A1W4XY44 A0A3B4YHB7 A0A1W4Y7P2 A0A3Q3IQ87 A0A195FIX9 Q3U0V9 T2HRB1 A0A3Q1HR28 Q3U1F2 A0A0R4IWQ3 Q8N6H6 Q792F9 A0A2J8NQC4 A0A1W4W0F9 E9PDS3 A0A3Q3Q209 A0A1A9XFD6 A0A1B0BAL8 A0A0F8B184 A0A2K5PV54 A0A1I8NPW8 A0A0N8AHQ2 A0A3P9NB79 F4W6F4 A0A3Q3SNW5 G1TR44 A0A151IQS9 A0A2K6LJ44 Q6NV53 A0A3B0J7L9 M3VY81 A0A3B0JT85 F6UVL3 A0A2K6PQH5 A0A091E739 F6TLC2 B4GBW0 A0A096M0L4 A0A0R4ICS1 A0A0P5BAH1
A0A212F4X5 A0A2W1B894 A0A2H1VUW8 Q86G86 A0A3S2NWJ7 A0A2H1V8F5 A0A0L7LUC9 A0A2J7QMA1 A0A3R7SKY0 A0A0A0PGH3 A0A087UCA8 A0A2Z5WPE6 A0A147BHT3 A0A034VIP4 A0A0K8V796 A0A0K8VM67 A0A0K8VIA7 A0A0A1XJR9 A0A0A1X2M1 Q16R90 Q16JC5 W8BH52 W8AK36 A0A182HAI7 A0A1I8N608 A0A293N3R2 A0A1Y1LAY1 A0A1W4WBX2 A0A3Q4N2Y5 A0A1Z5LDZ0 A0A3Q2VM08 F6R3Z0 A0A3B4YNS0 A0A0P4VVE3 A0A3B4U9I1 A0A1Y1LCJ3 A0A182Y859 A0A1A9VE28 A0A3Q3FQK3 A0A1Y1L8R1 I3JIG3 Q8BQ25 A0A3B4FXJ4 A0A195E483 A0A1A9WWH1 A0A151X2M7 A0A1B0FDC0 A0A1B0A219 A0A1W4XY44 A0A3B4YHB7 A0A1W4Y7P2 A0A3Q3IQ87 A0A195FIX9 Q3U0V9 T2HRB1 A0A3Q1HR28 Q3U1F2 A0A0R4IWQ3 Q8N6H6 Q792F9 A0A2J8NQC4 A0A1W4W0F9 E9PDS3 A0A3Q3Q209 A0A1A9XFD6 A0A1B0BAL8 A0A0F8B184 A0A2K5PV54 A0A1I8NPW8 A0A0N8AHQ2 A0A3P9NB79 F4W6F4 A0A3Q3SNW5 G1TR44 A0A151IQS9 A0A2K6LJ44 Q6NV53 A0A3B0J7L9 M3VY81 A0A3B0JT85 F6UVL3 A0A2K6PQH5 A0A091E739 F6TLC2 B4GBW0 A0A096M0L4 A0A0R4ICS1 A0A0P5BAH1
Pubmed
25064490
19121390
17868866
26354079
22118469
28756777
+ More
12974949 26227816 24982879 29652888 25348373 25830018 17510324 24495485 26483478 25315136 28004739 25186727 28528879 17495919 25244985 10349636 11042159 11076861 11217851 12466851 16141073 23594743 15489334 7772255 8756341 12040188 19144319 19468303 21183079 16641997 25835551 21719571 21993624 17975172 19892987 25362486 17994087
12974949 26227816 24982879 29652888 25348373 25830018 17510324 24495485 26483478 25315136 28004739 25186727 28528879 17495919 25244985 10349636 11042159 11076861 11217851 12466851 16141073 23594743 15489334 7772255 8756341 12040188 19144319 19468303 21183079 16641997 25835551 21719571 21993624 17975172 19892987 25362486 17994087
EMBL
KJ511850
AII79409.1
BABH01005253
BABH01005254
BABH01005255
BABH01005256
+ More
BABH01005257 DQ531846 ABF59520.1 NWSH01000832 PCG73951.1 KQ461194 KPJ06885.1 KQ459603 KPI92953.1 AGBW02010302 OWR48769.1 KZ150367 PZC71171.1 ODYU01004594 SOQ44635.1 AY237587 AAO85805.1 RSAL01000043 RVE50694.1 ODYU01001218 SOQ37130.1 JTDY01000080 KOB78989.1 NEVH01013215 PNF29696.1 QCYY01003118 ROT65225.1 KC715736 AHH32887.1 KK119179 KFM74997.1 LC114983 BBC20717.1 GEGO01005083 JAR90321.1 GAKP01017524 JAC41428.1 GDHF01017527 JAI34787.1 GDHF01012345 JAI39969.1 GDHF01026317 GDHF01014059 JAI25997.1 JAI38255.1 GBXI01014337 GBXI01003157 JAC99954.1 JAD11135.1 GBXI01009292 JAD05000.1 CH477721 EAT36913.1 CH478017 EAT34371.1 GAMC01017582 GAMC01017581 JAB88974.1 GAMC01017580 JAB88975.1 JXUM01005310 KQ560190 KXJ83915.1 GFWV01023037 MAA47764.1 GEZM01066073 JAV68207.1 GFJQ02001344 JAW05626.1 GDRN01105887 JAI57655.1 GEZM01062083 JAV70050.1 GEZM01062082 JAV70052.1 AERX01026760 AERX01026761 AK051676 BAC34715.1 KQ979657 KYN19896.1 KQ982580 KYQ54454.1 CCAG010020817 KQ981523 KYN40358.1 AK156517 BAE33742.1 AB850880 BAN82540.1 AK156015 BAE33546.1 CABZ01009769 FP102798 FP102862 BC030198 AAH30198.1 AL844590 AH003666 U34627 U34628 U34629 U34631 U34632 U34633 U34634 U34635 U34763 U34764 U34765 U34766 U34767 U34768 U34769 U34770 U34797 U34798 U34799 AF109136 CH466519 AAB09630.1 AAC95388.1 EDL27240.1 NBAG03000225 PNI73938.1 AC092055 AC093415 AP006240 JXJN01011053 KQ041982 KKF19882.1 GDIP01146321 JAJ77081.1 GL887707 EGI70289.1 AAGW02047530 AAGW02047531 AAGW02047532 AAGW02047533 AAGW02047534 AAGW02047535 AAGW02047536 AAGW02047537 KQ976755 KYN08585.1 BC068313 AAH68313.1 OUUW01000001 SPP75912.1 AANG04001816 SPP75911.1 KN122353 KFO30946.1 CH479181 EDW31339.1 AYCK01002109 GDIP01191525 JAJ31877.1
BABH01005257 DQ531846 ABF59520.1 NWSH01000832 PCG73951.1 KQ461194 KPJ06885.1 KQ459603 KPI92953.1 AGBW02010302 OWR48769.1 KZ150367 PZC71171.1 ODYU01004594 SOQ44635.1 AY237587 AAO85805.1 RSAL01000043 RVE50694.1 ODYU01001218 SOQ37130.1 JTDY01000080 KOB78989.1 NEVH01013215 PNF29696.1 QCYY01003118 ROT65225.1 KC715736 AHH32887.1 KK119179 KFM74997.1 LC114983 BBC20717.1 GEGO01005083 JAR90321.1 GAKP01017524 JAC41428.1 GDHF01017527 JAI34787.1 GDHF01012345 JAI39969.1 GDHF01026317 GDHF01014059 JAI25997.1 JAI38255.1 GBXI01014337 GBXI01003157 JAC99954.1 JAD11135.1 GBXI01009292 JAD05000.1 CH477721 EAT36913.1 CH478017 EAT34371.1 GAMC01017582 GAMC01017581 JAB88974.1 GAMC01017580 JAB88975.1 JXUM01005310 KQ560190 KXJ83915.1 GFWV01023037 MAA47764.1 GEZM01066073 JAV68207.1 GFJQ02001344 JAW05626.1 GDRN01105887 JAI57655.1 GEZM01062083 JAV70050.1 GEZM01062082 JAV70052.1 AERX01026760 AERX01026761 AK051676 BAC34715.1 KQ979657 KYN19896.1 KQ982580 KYQ54454.1 CCAG010020817 KQ981523 KYN40358.1 AK156517 BAE33742.1 AB850880 BAN82540.1 AK156015 BAE33546.1 CABZ01009769 FP102798 FP102862 BC030198 AAH30198.1 AL844590 AH003666 U34627 U34628 U34629 U34631 U34632 U34633 U34634 U34635 U34763 U34764 U34765 U34766 U34767 U34768 U34769 U34770 U34797 U34798 U34799 AF109136 CH466519 AAB09630.1 AAC95388.1 EDL27240.1 NBAG03000225 PNI73938.1 AC092055 AC093415 AP006240 JXJN01011053 KQ041982 KKF19882.1 GDIP01146321 JAJ77081.1 GL887707 EGI70289.1 AAGW02047530 AAGW02047531 AAGW02047532 AAGW02047533 AAGW02047534 AAGW02047535 AAGW02047536 AAGW02047537 KQ976755 KYN08585.1 BC068313 AAH68313.1 OUUW01000001 SPP75912.1 AANG04001816 SPP75911.1 KN122353 KFO30946.1 CH479181 EDW31339.1 AYCK01002109 GDIP01191525 JAJ31877.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000037510 UP000235965 UP000283509 UP000054359 UP000008820 UP000069940 UP000249989 UP000095301 UP000192221 UP000261580 UP000264840 UP000002280 UP000261360 UP000261420 UP000076408 UP000078200 UP000261660 UP000005207 UP000261460 UP000078492 UP000091820 UP000075809 UP000092444 UP000092445 UP000192224 UP000261600 UP000078541 UP000265040 UP000000437 UP000000589 UP000005640 UP000092443 UP000092460 UP000233040 UP000095300 UP000242638 UP000007755 UP000261640 UP000001811 UP000078542 UP000233180 UP000268350 UP000011712 UP000002281 UP000233200 UP000028990 UP000008744 UP000028760
UP000037510 UP000235965 UP000283509 UP000054359 UP000008820 UP000069940 UP000249989 UP000095301 UP000192221 UP000261580 UP000264840 UP000002280 UP000261360 UP000261420 UP000076408 UP000078200 UP000261660 UP000005207 UP000261460 UP000078492 UP000091820 UP000075809 UP000092444 UP000092445 UP000192224 UP000261600 UP000078541 UP000265040 UP000000437 UP000000589 UP000005640 UP000092443 UP000092460 UP000233040 UP000095300 UP000242638 UP000007755 UP000261640 UP000001811 UP000078542 UP000233180 UP000268350 UP000011712 UP000002281 UP000233200 UP000028990 UP000008744 UP000028760
Interpro
SUPFAM
SSF69179
SSF69179
Gene 3D
ProteinModelPortal
A0A076JQH6
H9JLQ2
Q1G0S5
A0A2A4JPM1
A0A194QPI3
A0A194PIS8
+ More
A0A212F4X5 A0A2W1B894 A0A2H1VUW8 Q86G86 A0A3S2NWJ7 A0A2H1V8F5 A0A0L7LUC9 A0A2J7QMA1 A0A3R7SKY0 A0A0A0PGH3 A0A087UCA8 A0A2Z5WPE6 A0A147BHT3 A0A034VIP4 A0A0K8V796 A0A0K8VM67 A0A0K8VIA7 A0A0A1XJR9 A0A0A1X2M1 Q16R90 Q16JC5 W8BH52 W8AK36 A0A182HAI7 A0A1I8N608 A0A293N3R2 A0A1Y1LAY1 A0A1W4WBX2 A0A3Q4N2Y5 A0A1Z5LDZ0 A0A3Q2VM08 F6R3Z0 A0A3B4YNS0 A0A0P4VVE3 A0A3B4U9I1 A0A1Y1LCJ3 A0A182Y859 A0A1A9VE28 A0A3Q3FQK3 A0A1Y1L8R1 I3JIG3 Q8BQ25 A0A3B4FXJ4 A0A195E483 A0A1A9WWH1 A0A151X2M7 A0A1B0FDC0 A0A1B0A219 A0A1W4XY44 A0A3B4YHB7 A0A1W4Y7P2 A0A3Q3IQ87 A0A195FIX9 Q3U0V9 T2HRB1 A0A3Q1HR28 Q3U1F2 A0A0R4IWQ3 Q8N6H6 Q792F9 A0A2J8NQC4 A0A1W4W0F9 E9PDS3 A0A3Q3Q209 A0A1A9XFD6 A0A1B0BAL8 A0A0F8B184 A0A2K5PV54 A0A1I8NPW8 A0A0N8AHQ2 A0A3P9NB79 F4W6F4 A0A3Q3SNW5 G1TR44 A0A151IQS9 A0A2K6LJ44 Q6NV53 A0A3B0J7L9 M3VY81 A0A3B0JT85 F6UVL3 A0A2K6PQH5 A0A091E739 F6TLC2 B4GBW0 A0A096M0L4 A0A0R4ICS1 A0A0P5BAH1
A0A212F4X5 A0A2W1B894 A0A2H1VUW8 Q86G86 A0A3S2NWJ7 A0A2H1V8F5 A0A0L7LUC9 A0A2J7QMA1 A0A3R7SKY0 A0A0A0PGH3 A0A087UCA8 A0A2Z5WPE6 A0A147BHT3 A0A034VIP4 A0A0K8V796 A0A0K8VM67 A0A0K8VIA7 A0A0A1XJR9 A0A0A1X2M1 Q16R90 Q16JC5 W8BH52 W8AK36 A0A182HAI7 A0A1I8N608 A0A293N3R2 A0A1Y1LAY1 A0A1W4WBX2 A0A3Q4N2Y5 A0A1Z5LDZ0 A0A3Q2VM08 F6R3Z0 A0A3B4YNS0 A0A0P4VVE3 A0A3B4U9I1 A0A1Y1LCJ3 A0A182Y859 A0A1A9VE28 A0A3Q3FQK3 A0A1Y1L8R1 I3JIG3 Q8BQ25 A0A3B4FXJ4 A0A195E483 A0A1A9WWH1 A0A151X2M7 A0A1B0FDC0 A0A1B0A219 A0A1W4XY44 A0A3B4YHB7 A0A1W4Y7P2 A0A3Q3IQ87 A0A195FIX9 Q3U0V9 T2HRB1 A0A3Q1HR28 Q3U1F2 A0A0R4IWQ3 Q8N6H6 Q792F9 A0A2J8NQC4 A0A1W4W0F9 E9PDS3 A0A3Q3Q209 A0A1A9XFD6 A0A1B0BAL8 A0A0F8B184 A0A2K5PV54 A0A1I8NPW8 A0A0N8AHQ2 A0A3P9NB79 F4W6F4 A0A3Q3SNW5 G1TR44 A0A151IQS9 A0A2K6LJ44 Q6NV53 A0A3B0J7L9 M3VY81 A0A3B0JT85 F6UVL3 A0A2K6PQH5 A0A091E739 F6TLC2 B4GBW0 A0A096M0L4 A0A0R4ICS1 A0A0P5BAH1
PDB
4IRZ
E-value=8.23729e-42,
Score=430
Ontologies
GO
GO:0008305
GO:0007229
GO:0007155
GO:0016021
GO:0016020
GO:0019960
GO:1904646
GO:0050839
GO:0043113
GO:2000353
GO:0035987
GO:0009986
GO:0050901
GO:1905564
GO:0034446
GO:0140039
GO:0003366
GO:0034113
GO:0090074
GO:0034669
GO:0033631
GO:1903238
GO:2000406
GO:0070062
GO:0060385
GO:1990771
GO:0030426
GO:0071345
GO:1990138
GO:0050904
GO:1990405
GO:0043025
GO:0001968
GO:0009925
GO:0030593
GO:0004965
GO:0030286
GO:0005515
GO:0004319
GO:0004320
GO:0016295
GO:0016296
Topology
Subcellular location
Membrane
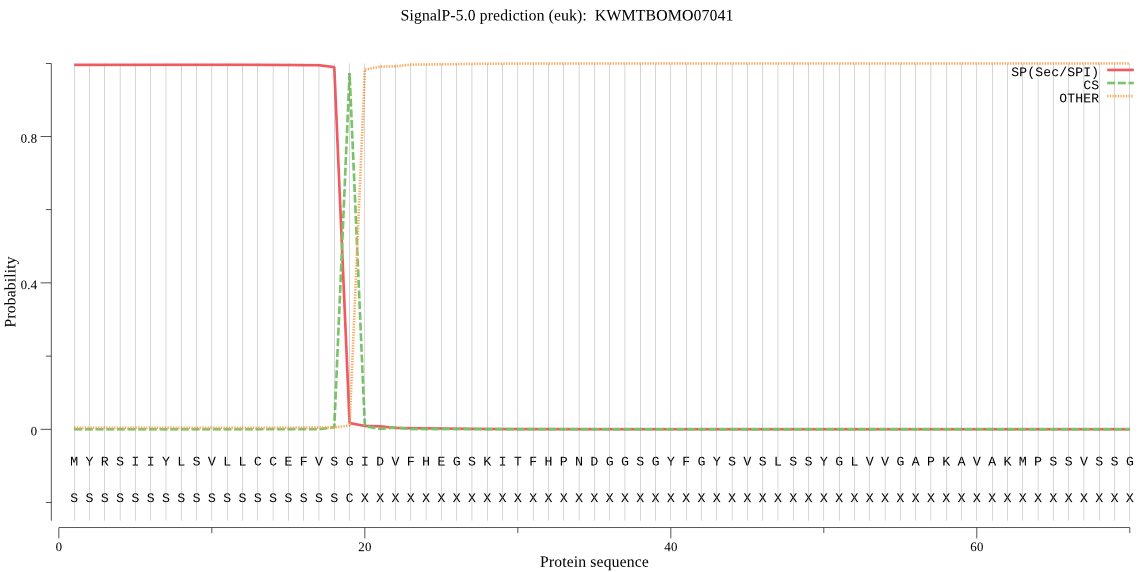
SignalP
Position: 1 - 19,
Likelihood: 0.995706
Length:
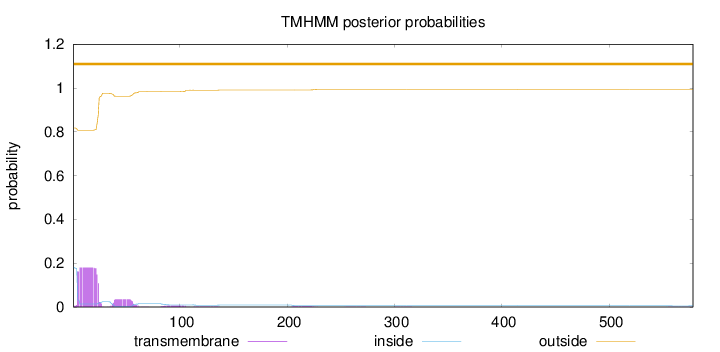
578
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.48436
Exp number, first 60 AAs:
4.21762
Total prob of N-in:
0.18292
outside
1 - 578
Population Genetic Test Statistics
Pi
193.087042
Theta
160.852615
Tajima's D
0.375593
CLR
0.012905
CSRT
0.471126443677816
Interpretation
Uncertain
