Gene
KWMTBOMO07029
Pre Gene Modal
BGIBMGA010460
Annotation
PREDICTED:_gamma-aminobutyric_acid_type_B_receptor_subunit_2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.343
Sequence
CDS
ATGTACGAGCCAAATATATCAAACGAAACAAATATATTGAATGAAAATTTTAATGACTTAACGAACAACACCAATAATGAGTTTTCAGAAGACGTTCACGAATTTATTGGTACAGTGAAACTGGAAAATATAACTAAAGATCTAATCACTGATTTTAATGCAACAATATCTGAATCTAATGAATCAGATATTGGTGACGATTTAAAAGCTATAGAAATAGGTGTACTTAATATAGAAAATAAAGATAATTCAATGGAAAAACTCAGAGTCGATGACGTAACGAGAACTAAAATATTTTACGATTTTTCGACCACAGTCAACAAGAAAGATGACAGAAATTTAATTACTAATGAAACGTTTACAGTAAAATTCAATTTAGATAAAAACAAAACGACTGAGGCCAAAATACGTGACTTAAAGAATTATCCGAGCGGATCTTTTCATCCTATCCTAAATCCTCCCGCTCAAATTACGGACGTGAATACTTCTAGTTCAACTTTTATAAACGAATTACCAAGAGCCACAACCAGCCGCGTCGTGTCTATCCTCGGTCTCTTCGAGTTGTCTGTGGGAGATACACCAAGAGTGGACGGAGCATCAGAGCTCGCTGCTGCCAAGCTCGCAGTGGAACATGTCAACAAAAGGAACCTACTCCCTGGGTATAAGTTGCATCTCATCACCAATGACACAAAGTGCGATCCAGGTGTGGGCGTGGATAGGTTCTTTCACGCCCTCTACACGGAGCGGGAGTCAAGAATGGTCATGCTGCTGGGGACTGCTTGTTCTGAAGTCACGGAGAGCATCGCGAAAATCGTACCCTATTGGAACATTGTACAGGTGTCCTTCGGTTCCACGTCACCAGCTCTGAGTGATAGATCAGAATTTCCCCTCTTCTGCCGCACCGTCGCGCCTGATTCCTCACATAACCCCGCACGGATAGCTTTCATCAGACAATTCGGATGGGACACGGTGACGGCTTTCTCGCAGAACGAGGAGGTTTACTCGTTGGCTGTGAACGAGCTGGTCACGCAACTAGAAGCCGCCAATATCACCTGTGCCGCCTCCATCACCTTCGCGGAGTCGGACTTCAAGGAGCAACTGCAACAGTTAAAGGAACTAGATACAAGAATTATAATCGGAAGCTTTTCTCAGGAGATGATACCGAAAATATTTTGCGAGGCTTTCCGGTTGGGGATGTTTGGAGCGGAATATGCCTGGCTTGTTGCCGGCATGCCGCCTCGCATTAGAGGCACCGCGCCTTGCGCCAGGTCTCAGCTGGCCAGGGCCTTAGAAGGCCTCGTCACAGTCACAGCTTATAGAGGAATCGTGGGAGACGTGGCTTCATTCTCTGGGTTGACAAACCAAATGTTTCACCGTGAGATGGAAGCCGTAGGAGTACCAGTCGCTCCGTACGCCCCACAAACCTACGACGCAGTCTGGGTCATAGCACTAGCACTAAGCAGAGCTGAACAGCAATGGAGTCAGGGTATCAATGGAACGCCTACGAACAATAAACTGGCCCTTGACCATTTCGACTACGACCGGAAGGACATGGCCGAGGACTTCCTCAATCAGTTGGAGAGTTTGAGTTTTTTGGGTGTTTCTGGACCTGTATCGTTCAATGGCGCAGACAGAATCGGTACATCAGCGTTTTATCAAATTCAAGGTGGTCGTCCAAAAACCGTTGCATTATACACCCATGGCAGAGAAATCTCGTGTACTGACTGCGCCAAGCCACAATGGGCTGGTGGCATACCCGCGGCCAGACGGGTACTCGTTCTAAGGGTCGATTCCGTGTGGGCTCCCGCTAGACTGGCGGTGGCTACATTGGCAGCAGCTGGCGTGGCTCTTGCGCTGGCATTTCTGGTCTTTAATTTACACTACAGTAAAAGAAGATCAATAAAACTCTCATCTCCGAGACTGAACAACATGACTCTGATTGGTTGCGTGCTAGTATATACTGCTGTTGCGTTATTGGGCGTCGACAATGCCACTTTACCATCATACGTGCCCTTTTCTGCCATGTGTACTGGTAGAGTGTACATTCTGTCGGCTGGTTTTTCATTGGCTTTTGGTTCGATGTTTGCCAAAACGTACAGAGTTCATCGGATATTTGTAAGTAACAGAAGTGGTGTCTGCAAAACGAAGCTACTCCAAGATACGCCTCTCATATCCCTGGTCTGCGCTTTACTTGTGATCGACGGCATGATCGTCACTCTCTGGACCATCCTAGATCCAATGGAGAGACACCTGAAGAACTTGACGATTGAGATCAGCGCTAACGATAGAAGCGTGATTTATCAACCTCAGCTAGAAGTCTGCAAGTCTCAGAACACAACGGGCTGGCTTTGCGCTTTATACGCTTACAAGGGCCTCCTTCTCATAGTGGGGGTGTACATGGCTTGGGAGACCAGAAACGTTAAGATTTCTGCCCTAAACGACTCGAAATACATCGGAATCAGTGTTTATTCTGTTGTGATCACCAGCGCAAGTGTTGTTGTTATAGGAACCATAATTTCCGAAAGAGCCACTTTGGCTTACATCAGTATAACTAGTTTGATCCTTATTACAACGACATCAACGCTCTGTTTGCTGTTCTTGCCGAAAATTGTCGCAATTAGAAGGAATATTGAAGAAGACCCCGTCATACAAAGCATGGGCCTGAGGCTGGAGTGTAAAACCAGAAGGTTCGTTACGGACGAAACCCAAGAACTACATTTCAGGATAGAAATACAGAATAAAGTGTTCAGGAGAGAAGTGGCGGCGCTGGACAAAGAAATCGGAAGACTTGAGAAACTGCTATCAGAGCCGCTAGACTTCACTGGGTCAAACGCCTCGATATCCTTGCAGAGACGCTTGGAGCTCAACACAGTCGAAGAGGAAGGTAATCTACGTTACGACGATCGACGTACTCCAAGCATCGGCGGTGGCCTACCCATGCTCCTGCTGTCAGTTCTACCGCCAGTCATACCCCGAGCAAGCTGGCCCTCAGCCGAAGCCTATCGCGGCAGGAACTCCGTCAGTTTCAGCTCGCAACCTAAGCTCGACCAAAGGAAATCGCATCCAGCGATCGACTTATACAATTTGTGTCTCAATAAACGAGAACAGCCCGGCTTACTCAGAAGACTGAAAAGCTTCTTCGGCAGTAGACCTTCCAGTAGAAAGGCGTCTGCCATATCAATAGCCGACCCGACAAGTCAAAGCATCGCGGCAGCTTTGAAAATGCACGTCGGAATGATCGCAGGATTAGTTCCGGGAACCAGAAAGCACTCAATGGTGTTATCCTGCAACACGCTACACGTGCCGAACCCCGAGATGAAACTGAGAAGAGAGTCTTACGCGAAAAGCGGCCCTATCATTCGGGTAACCGATGACGAGCCGTCCTGCTCAAGTTACATTCCGGAGAACAGCGTACACATAGGGCCCCAGAAATATATTGCTGAGCACGAGACCAAAGTAAACTTTGTATTACCACCGACACATCAAAGACCAATGTCCCACCAGAGCTCGTATGAAAAAATCAAGGGCTCTCCAAGGTTTCCTCATAGGATAACTCCTACAGCTAGCAGCTTGACCAATAGTCTTCTAGATACAAGAAAAACCAGTGCCGGTAGCATTTTGAGCGTGTCTGGCAAGGCCTTTGATGAGCAGCGAAGGTTTAGTCATCAGAACGTCAGCCATCAAGAAGATAGAGGAAAGACCCTACTAGAAAGCAGATCGAAGTCTACAGAAGACGCAATGCCTGGGCCTTCGACGAGGAATGATTAA
Protein
MYEPNISNETNILNENFNDLTNNTNNEFSEDVHEFIGTVKLENITKDLITDFNATISESNESDIGDDLKAIEIGVLNIENKDNSMEKLRVDDVTRTKIFYDFSTTVNKKDDRNLITNETFTVKFNLDKNKTTEAKIRDLKNYPSGSFHPILNPPAQITDVNTSSSTFINELPRATTSRVVSILGLFELSVGDTPRVDGASELAAAKLAVEHVNKRNLLPGYKLHLITNDTKCDPGVGVDRFFHALYTERESRMVMLLGTACSEVTESIAKIVPYWNIVQVSFGSTSPALSDRSEFPLFCRTVAPDSSHNPARIAFIRQFGWDTVTAFSQNEEVYSLAVNELVTQLEAANITCAASITFAESDFKEQLQQLKELDTRIIIGSFSQEMIPKIFCEAFRLGMFGAEYAWLVAGMPPRIRGTAPCARSQLARALEGLVTVTAYRGIVGDVASFSGLTNQMFHREMEAVGVPVAPYAPQTYDAVWVIALALSRAEQQWSQGINGTPTNNKLALDHFDYDRKDMAEDFLNQLESLSFLGVSGPVSFNGADRIGTSAFYQIQGGRPKTVALYTHGREISCTDCAKPQWAGGIPAARRVLVLRVDSVWAPARLAVATLAAAGVALALAFLVFNLHYSKRRSIKLSSPRLNNMTLIGCVLVYTAVALLGVDNATLPSYVPFSAMCTGRVYILSAGFSLAFGSMFAKTYRVHRIFVSNRSGVCKTKLLQDTPLISLVCALLVIDGMIVTLWTILDPMERHLKNLTIEISANDRSVIYQPQLEVCKSQNTTGWLCALYAYKGLLLIVGVYMAWETRNVKISALNDSKYIGISVYSVVITSASVVVIGTIISERATLAYISITSLILITTTSTLCLLFLPKIVAIRRNIEEDPVIQSMGLRLECKTRRFVTDETQELHFRIEIQNKVFRREVAALDKEIGRLEKLLSEPLDFTGSNASISLQRRLELNTVEEEGNLRYDDRRTPSIGGGLPMLLLSVLPPVIPRASWPSAEAYRGRNSVSFSSQPKLDQRKSHPAIDLYNLCLNKREQPGLLRRLKSFFGSRPSSRKASAISIADPTSQSIAAALKMHVGMIAGLVPGTRKHSMVLSCNTLHVPNPEMKLRRESYAKSGPIIRVTDDEPSCSSYIPENSVHIGPQKYIAEHETKVNFVLPPTHQRPMSHQSSYEKIKGSPRFPHRITPTASSLTNSLLDTRKTSAGSILSVSGKAFDEQRRFSHQNVSHQEDRGKTLLESRSKSTEDAMPGPSTRND
Summary
Similarity
Belongs to the G-protein coupled receptor 3 family.
Uniprot
H9JLQ8
A0A2H1V0S0
A0A3S2LEA5
A0A194QTK8
A0A194PIT2
A0A336LRP5
+ More
A0A067RF46 A0A154PTY8 B4JDS6 B4LTP6 A0A0C9QHY7 B4P2W6 B3MLQ9 B3N7T8 A0A0J9QTA5 A0A3B0JBL7 Q9BML5 B4KF86 Q9VPS7 Q29LM3 A0A3L8DVA8 B4ID07 A0A1W4UQ97 A0A088AN28 A0A2A3EI96 B4N0S2 A0A158NYX1 A0A195BUN8 A0A0L7RDY1 B4Q6J0 A0A1B0CFC1 A0A0M8ZYE3 A0A026WBP1 A0A151WQG7 A0A151J159 A0A182GGK9 A0A1J1I188 A0A182Y5U1 A0A182V1P6 A0A232F408 A0A182X7M6 A0A182F168 A0A1S4H1D5 Q171A7 A0A182IE41 W5JV31 A0A182TPY1 A0A084VPF7 A0A1S4J7W5 B0W5U1 A0A195FTL5 Q7PFP4 A0A195D0T9 A0A0N8NZ11 A0A182Q0P0 A0A182NEE9 A0A182P375 A0A182RGF3 A0A182VUZ2 A0A182KGY4 A0A182LUW1 B4GPQ7 A0A182JFC5 A0A2J7RC43 D2A100 T1HAH4
A0A067RF46 A0A154PTY8 B4JDS6 B4LTP6 A0A0C9QHY7 B4P2W6 B3MLQ9 B3N7T8 A0A0J9QTA5 A0A3B0JBL7 Q9BML5 B4KF86 Q9VPS7 Q29LM3 A0A3L8DVA8 B4ID07 A0A1W4UQ97 A0A088AN28 A0A2A3EI96 B4N0S2 A0A158NYX1 A0A195BUN8 A0A0L7RDY1 B4Q6J0 A0A1B0CFC1 A0A0M8ZYE3 A0A026WBP1 A0A151WQG7 A0A151J159 A0A182GGK9 A0A1J1I188 A0A182Y5U1 A0A182V1P6 A0A232F408 A0A182X7M6 A0A182F168 A0A1S4H1D5 Q171A7 A0A182IE41 W5JV31 A0A182TPY1 A0A084VPF7 A0A1S4J7W5 B0W5U1 A0A195FTL5 Q7PFP4 A0A195D0T9 A0A0N8NZ11 A0A182Q0P0 A0A182NEE9 A0A182P375 A0A182RGF3 A0A182VUZ2 A0A182KGY4 A0A182LUW1 B4GPQ7 A0A182JFC5 A0A2J7RC43 D2A100 T1HAH4
Pubmed
EMBL
BABH01005228
BABH01005229
BABH01005230
BABH01005231
ODYU01000155
SOQ34445.1
+ More
RSAL01000028 RVE51879.1 KQ461194 KPJ06881.1 KQ459603 KPI92958.1 UFQT01000134 SSX20630.1 KK852674 KDR18733.1 KQ435102 KZC14670.1 CH916368 EDW03446.1 CH940649 EDW65019.1 GBYB01014278 JAG84045.1 CM000157 EDW87173.1 CH902620 EDV30780.1 CH954177 EDV57264.1 CM002910 KMY87357.1 KMY87359.1 OUUW01000004 SPP79744.1 AF318274 AAK13422.1 CH933807 EDW13069.2 KRG03565.1 KRG03566.1 KRG03567.1 KRG03568.1 AE014134 AAF51465.2 AAN10484.2 AFH03503.1 AHN54069.1 CH379060 EAL34022.3 QOIP01000003 RLU24420.1 CH480829 EDW45433.1 KZ288245 PBC31194.1 CH963920 EDW77685.2 ADTU01004332 ADTU01004333 ADTU01004334 ADTU01004335 ADTU01004336 ADTU01004337 ADTU01004338 ADTU01004339 ADTU01004340 ADTU01004341 KQ976407 KYM91462.1 KQ414613 KOC69015.1 CM000361 EDX03256.1 AJWK01009929 AJWK01009930 AJWK01009931 AJWK01009932 AJWK01009933 AJWK01009934 KQ435824 KOX72109.1 KK107295 EZA53061.1 KQ982821 KYQ50152.1 KQ980571 KYN15529.1 JXUM01061926 KQ562175 KXJ76496.1 CVRI01000038 CRK94045.1 NNAY01000996 OXU25566.1 AAAB01008839 CH477460 EAT40585.2 APCN01004559 ADMH02000366 ETN66815.1 ATLV01015001 KE524999 KFB39851.1 DS231844 EDS35744.1 KQ981276 KYN43632.1 EAA45235.2 KQ977004 KYN06530.1 KPU73028.1 KPU73029.1 AXCN02000859 AXCM01002236 CH479187 EDW39579.1 NEVH01005887 PNF38414.1 KQ971338 EFA01604.1 ACPB03017385
RSAL01000028 RVE51879.1 KQ461194 KPJ06881.1 KQ459603 KPI92958.1 UFQT01000134 SSX20630.1 KK852674 KDR18733.1 KQ435102 KZC14670.1 CH916368 EDW03446.1 CH940649 EDW65019.1 GBYB01014278 JAG84045.1 CM000157 EDW87173.1 CH902620 EDV30780.1 CH954177 EDV57264.1 CM002910 KMY87357.1 KMY87359.1 OUUW01000004 SPP79744.1 AF318274 AAK13422.1 CH933807 EDW13069.2 KRG03565.1 KRG03566.1 KRG03567.1 KRG03568.1 AE014134 AAF51465.2 AAN10484.2 AFH03503.1 AHN54069.1 CH379060 EAL34022.3 QOIP01000003 RLU24420.1 CH480829 EDW45433.1 KZ288245 PBC31194.1 CH963920 EDW77685.2 ADTU01004332 ADTU01004333 ADTU01004334 ADTU01004335 ADTU01004336 ADTU01004337 ADTU01004338 ADTU01004339 ADTU01004340 ADTU01004341 KQ976407 KYM91462.1 KQ414613 KOC69015.1 CM000361 EDX03256.1 AJWK01009929 AJWK01009930 AJWK01009931 AJWK01009932 AJWK01009933 AJWK01009934 KQ435824 KOX72109.1 KK107295 EZA53061.1 KQ982821 KYQ50152.1 KQ980571 KYN15529.1 JXUM01061926 KQ562175 KXJ76496.1 CVRI01000038 CRK94045.1 NNAY01000996 OXU25566.1 AAAB01008839 CH477460 EAT40585.2 APCN01004559 ADMH02000366 ETN66815.1 ATLV01015001 KE524999 KFB39851.1 DS231844 EDS35744.1 KQ981276 KYN43632.1 EAA45235.2 KQ977004 KYN06530.1 KPU73028.1 KPU73029.1 AXCN02000859 AXCM01002236 CH479187 EDW39579.1 NEVH01005887 PNF38414.1 KQ971338 EFA01604.1 ACPB03017385
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000027135
UP000076502
+ More
UP000001070 UP000008792 UP000002282 UP000007801 UP000008711 UP000268350 UP000009192 UP000000803 UP000001819 UP000279307 UP000001292 UP000192221 UP000005203 UP000242457 UP000007798 UP000005205 UP000078540 UP000053825 UP000000304 UP000092461 UP000053105 UP000053097 UP000075809 UP000078492 UP000069940 UP000249989 UP000183832 UP000076408 UP000075903 UP000215335 UP000076407 UP000069272 UP000008820 UP000075840 UP000000673 UP000075902 UP000030765 UP000002320 UP000078541 UP000007062 UP000078542 UP000075886 UP000075884 UP000075885 UP000075900 UP000075920 UP000075881 UP000075883 UP000008744 UP000075880 UP000235965 UP000007266 UP000015103
UP000001070 UP000008792 UP000002282 UP000007801 UP000008711 UP000268350 UP000009192 UP000000803 UP000001819 UP000279307 UP000001292 UP000192221 UP000005203 UP000242457 UP000007798 UP000005205 UP000078540 UP000053825 UP000000304 UP000092461 UP000053105 UP000053097 UP000075809 UP000078492 UP000069940 UP000249989 UP000183832 UP000076408 UP000075903 UP000215335 UP000076407 UP000069272 UP000008820 UP000075840 UP000000673 UP000075902 UP000030765 UP000002320 UP000078541 UP000007062 UP000078542 UP000075886 UP000075884 UP000075885 UP000075900 UP000075920 UP000075881 UP000075883 UP000008744 UP000075880 UP000235965 UP000007266 UP000015103
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
H9JLQ8
A0A2H1V0S0
A0A3S2LEA5
A0A194QTK8
A0A194PIT2
A0A336LRP5
+ More
A0A067RF46 A0A154PTY8 B4JDS6 B4LTP6 A0A0C9QHY7 B4P2W6 B3MLQ9 B3N7T8 A0A0J9QTA5 A0A3B0JBL7 Q9BML5 B4KF86 Q9VPS7 Q29LM3 A0A3L8DVA8 B4ID07 A0A1W4UQ97 A0A088AN28 A0A2A3EI96 B4N0S2 A0A158NYX1 A0A195BUN8 A0A0L7RDY1 B4Q6J0 A0A1B0CFC1 A0A0M8ZYE3 A0A026WBP1 A0A151WQG7 A0A151J159 A0A182GGK9 A0A1J1I188 A0A182Y5U1 A0A182V1P6 A0A232F408 A0A182X7M6 A0A182F168 A0A1S4H1D5 Q171A7 A0A182IE41 W5JV31 A0A182TPY1 A0A084VPF7 A0A1S4J7W5 B0W5U1 A0A195FTL5 Q7PFP4 A0A195D0T9 A0A0N8NZ11 A0A182Q0P0 A0A182NEE9 A0A182P375 A0A182RGF3 A0A182VUZ2 A0A182KGY4 A0A182LUW1 B4GPQ7 A0A182JFC5 A0A2J7RC43 D2A100 T1HAH4
A0A067RF46 A0A154PTY8 B4JDS6 B4LTP6 A0A0C9QHY7 B4P2W6 B3MLQ9 B3N7T8 A0A0J9QTA5 A0A3B0JBL7 Q9BML5 B4KF86 Q9VPS7 Q29LM3 A0A3L8DVA8 B4ID07 A0A1W4UQ97 A0A088AN28 A0A2A3EI96 B4N0S2 A0A158NYX1 A0A195BUN8 A0A0L7RDY1 B4Q6J0 A0A1B0CFC1 A0A0M8ZYE3 A0A026WBP1 A0A151WQG7 A0A151J159 A0A182GGK9 A0A1J1I188 A0A182Y5U1 A0A182V1P6 A0A232F408 A0A182X7M6 A0A182F168 A0A1S4H1D5 Q171A7 A0A182IE41 W5JV31 A0A182TPY1 A0A084VPF7 A0A1S4J7W5 B0W5U1 A0A195FTL5 Q7PFP4 A0A195D0T9 A0A0N8NZ11 A0A182Q0P0 A0A182NEE9 A0A182P375 A0A182RGF3 A0A182VUZ2 A0A182KGY4 A0A182LUW1 B4GPQ7 A0A182JFC5 A0A2J7RC43 D2A100 T1HAH4
PDB
4MS4
E-value=8.13954e-50,
Score=502
Ontologies
GO
PANTHER
Topology
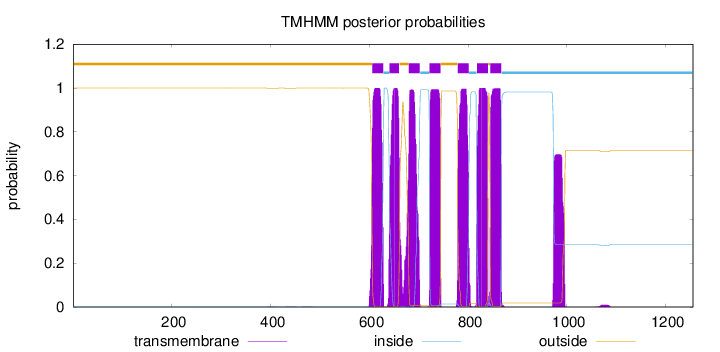
Length:
1255
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
169.27548
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00038
outside
1 - 605
TMhelix
606 - 628
inside
629 - 640
TMhelix
641 - 660
outside
661 - 679
TMhelix
680 - 702
inside
703 - 721
TMhelix
722 - 744
outside
745 - 778
TMhelix
779 - 801
inside
802 - 817
TMhelix
818 - 840
outside
841 - 844
TMhelix
845 - 867
inside
868 - 1255
Population Genetic Test Statistics
Pi
25.832165
Theta
213.453592
Tajima's D
-1.508289
CLR
0.635174
CSRT
0.0590970451477426
Interpretation
Uncertain
