Gene
KWMTBOMO07028
Pre Gene Modal
BGIBMGA010510
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_DDX28_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.112 Nuclear Reliability : 1.691
Sequence
CDS
ATGTCTAAATTACGTCAATTAATTAGCACATCTTATAGTCGTTATTATAGCCTCCAGCAGGCTAAGAAAAAATTACCTATTATAACATGCAAGCGACCGGAATTCAATCACTACGAAGGACAGAGTTATAGCAAGTATGAAGGTATAAAACTAGCTTCGCAGGGGTGGCTACACAATAAATCAAAGGGAGACTATTTCATCATACACGGGAATGCAAATAAAAAAGAAGAAACACCCGTGTATAGGAAGACTTTTGAAGATATCGGCTTGAAAGATAATTTGGTTAAAGTTGTCAAAGATTTAGGGTTTACGCTGCCCACGGCCATACAAACAAAGGCTGTACCCGCCATTCTCAATGGACACAATACGGTAATCACAGCGGAGACGGGCTGCGGGAAGACCTTAGCGTATTTGTTACCTATAATTCAACATATACTGGAATGGAAGCCAAGTATTCAAGAAGAATTTAATAGTCCATTGGCTGTTGTTATTACACCGAACAGAGAACTAGCTCTACAAATAGGAGAAGTGGCTCAAACAATAGCACAAAGCATAAACATAAATGTCACGACTTTAATAGGAGGCAAAACGAAAAAAAAAATGTTAAACCCTCCAATTGAGCACAGTGACATATTAATCACAACGCTCGGAGCTTACAGTAAACTAGTCACAACTGGAATATACAAGATTCACAATGTGCATCATATTGTGCTGGATGAAGCAGATTCCCTTTTGGATGACACATTTATTGATAAAGTCACATATCTGTTGAAGAAATTTTCGATACAATATAAAGTTGATATTCAAGTCCCTCCCACAGGTTGTCAGTTGACTTTAGTTAGTGCAACACTACCTAGGGACCTTCCGGAAACTGTCAATAGTTTTGTAGCGGCTGAATCACTGAAAATGGTTAGAACTTCAAATATACATCGGATATTACCACATGTGCCGCAGAAATTCTTACGTCTCGGGAAGGCGCACAAGCCCGCGGAACTCCTCAAACTTGTCCAAGCGGATCAGAACGAGAGGAGACCGGTCATGATCTTCAGCAATCAGACGTCTGGCTGCGACTTCGTGTCCATGTTCCTGAACGAGAACAACGTTGAATGCATCAACATCAACGGCCAGATGCCGGTACAGCTGAAGTTGGGGAAGTTTGAAATGTTCAGATCCGGGAAAGTTAATGTCATTTCGTGTACTGATATCGTTTCCCGAGGATTGGACACTACTAGGACGCACCACGTAATTAATTACGATTTCCCTCTGCACACGTCTGACTACATCCACCGGTGCGGGCGCGCGGGCCGGCTGGGCTCCCCGCCCGGATCCCACGTCACAAGCTTCGTGGCCTGGCCCCGCGAAGTCGAGCTGGTCCAGAAGATCGAGACCTCGGTCAGGACAAACTCTGACCTGCCCCACGTTAACGCCAACATCAAGCGGGTCATACGAGACAGGATTGTCAGGATGAGCGGGGCTCTTGCTAGGGCAAGTTGA
Protein
MSKLRQLISTSYSRYYSLQQAKKKLPIITCKRPEFNHYEGQSYSKYEGIKLASQGWLHNKSKGDYFIIHGNANKKEETPVYRKTFEDIGLKDNLVKVVKDLGFTLPTAIQTKAVPAILNGHNTVITAETGCGKTLAYLLPIIQHILEWKPSIQEEFNSPLAVVITPNRELALQIGEVAQTIAQSININVTTLIGGKTKKKMLNPPIEHSDILITTLGAYSKLVTTGIYKIHNVHHIVLDEADSLLDDTFIDKVTYLLKKFSIQYKVDIQVPPTGCQLTLVSATLPRDLPETVNSFVAAESLKMVRTSNIHRILPHVPQKFLRLGKAHKPAELLKLVQADQNERRPVMIFSNQTSGCDFVSMFLNENNVECININGQMPVQLKLGKFEMFRSGKVNVISCTDIVSRGLDTTRTHHVINYDFPLHTSDYIHRCGRAGRLGSPPGSHVTSFVAWPREVELVQKIETSVRTNSDLPHVNANIKRVIRDRIVRMSGALARAS
Summary
Uniprot
H9JLV8
A0A2W1BJ26
A0A2A4K814
A0A2H1V0T2
A0A3S2NY02
A0A194PJQ1
+ More
A0A194QPH8 A0A212EH25 A0A0L7L6I0 S4PBL6 A0A067RK13 A0A2J7PS80 A0A1B6EFR9 A0A1B6JBV5 T1IFZ3 A0A1W4V3N9 A0A224XIG1 A0A3R7QIC6 A0A0A9Z2K7 B4Q6J6 A0A1B6M4H1 B3N885 A0A1B6KUR4 B4ID15 B3MAI3 Q9VPT3 A0A3B0K0J5 B4P2X4 A0A1Q3EYR4 A0A034WBB6 W8B0D8 B4G9K2 A0A0K8V8M1 B0XJF6 W5JE88 A0A1J1ILH0 B0X2Y7 A0A1I8Q410 Q29M92 Q17CQ0 B4N0T5 A0A182H4Z1 A0A0P6HED8 A0A0P5CH29 A0A0P5T140 A0A0P5SYF6 B4JDT1 A0A2J7PS79 E9HSC9 A0A0M4E8R6 A0A084VAK2 B4LTQ1 A0A182PT25 A0A182YHI1 A0A1I8MM25 A0A2S2NQZ6 A0A182SY94 A0A1B0GQP0 A0A2H8TMY3 A0A1L8DRL1 A0A182VWM6 A0A182LVC9 A0A1B0C8R6 J9JJJ7 A0A182Q8K6 A0A182HJ83 A0A1B0C103 A0A182LCF1 A0A182XE40 A0A182TYQ9 A0A182UPG5 B4KF80 A0A0V0GBP4 D3TPD0 A0A0L0C8U0 A0A1B0A7M7 A0A158NZ67 A0A232FG16 K7IQP8 A0A1A9YIR9 A0A0T6AUP4 A0A0P5YDA7 A0A182N510 A0A195AYK6 A0A195CW58 A0A1A9UNS1 Q7QM14 A0A2S2Q052 E9IUV1 A0A195F6U8 A0A151WQ97 A0A182FKU6 A0A026WQB9 A0A0P5EDW2 A0A182RC47 F4X348 A0A151J3Z5 A0A154PCU6 A0A226EW59 A0A0N8EDW2
A0A194QPH8 A0A212EH25 A0A0L7L6I0 S4PBL6 A0A067RK13 A0A2J7PS80 A0A1B6EFR9 A0A1B6JBV5 T1IFZ3 A0A1W4V3N9 A0A224XIG1 A0A3R7QIC6 A0A0A9Z2K7 B4Q6J6 A0A1B6M4H1 B3N885 A0A1B6KUR4 B4ID15 B3MAI3 Q9VPT3 A0A3B0K0J5 B4P2X4 A0A1Q3EYR4 A0A034WBB6 W8B0D8 B4G9K2 A0A0K8V8M1 B0XJF6 W5JE88 A0A1J1ILH0 B0X2Y7 A0A1I8Q410 Q29M92 Q17CQ0 B4N0T5 A0A182H4Z1 A0A0P6HED8 A0A0P5CH29 A0A0P5T140 A0A0P5SYF6 B4JDT1 A0A2J7PS79 E9HSC9 A0A0M4E8R6 A0A084VAK2 B4LTQ1 A0A182PT25 A0A182YHI1 A0A1I8MM25 A0A2S2NQZ6 A0A182SY94 A0A1B0GQP0 A0A2H8TMY3 A0A1L8DRL1 A0A182VWM6 A0A182LVC9 A0A1B0C8R6 J9JJJ7 A0A182Q8K6 A0A182HJ83 A0A1B0C103 A0A182LCF1 A0A182XE40 A0A182TYQ9 A0A182UPG5 B4KF80 A0A0V0GBP4 D3TPD0 A0A0L0C8U0 A0A1B0A7M7 A0A158NZ67 A0A232FG16 K7IQP8 A0A1A9YIR9 A0A0T6AUP4 A0A0P5YDA7 A0A182N510 A0A195AYK6 A0A195CW58 A0A1A9UNS1 Q7QM14 A0A2S2Q052 E9IUV1 A0A195F6U8 A0A151WQ97 A0A182FKU6 A0A026WQB9 A0A0P5EDW2 A0A182RC47 F4X348 A0A151J3Z5 A0A154PCU6 A0A226EW59 A0A0N8EDW2
Pubmed
19121390
28756777
26354079
22118469
26227816
23622113
+ More
24845553 25401762 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 24495485 20920257 23761445 15632085 17510324 26483478 21292972 24438588 25244985 25315136 20966253 20353571 26108605 21347285 28648823 20075255 9087549 12364791 14747013 17210077 21282665 24508170 21719571
24845553 25401762 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 24495485 20920257 23761445 15632085 17510324 26483478 21292972 24438588 25244985 25315136 20966253 20353571 26108605 21347285 28648823 20075255 9087549 12364791 14747013 17210077 21282665 24508170 21719571
EMBL
BABH01005227
KZ150274
PZC71663.1
NWSH01000039
PCG80371.1
ODYU01000155
+ More
SOQ34447.1 RSAL01000028 RVE51878.1 KQ459603 KPI92959.1 KQ461194 KPJ06880.1 AGBW02014975 OWR40789.1 JTDY01002689 KOB70916.1 GAIX01004561 JAA87999.1 KK852424 KDR24181.1 NEVH01021938 PNF19192.1 GEDC01000520 JAS36778.1 GECU01010985 JAS96721.1 ACPB03004178 GFTR01006818 JAW09608.1 QCYY01001184 ROT79860.1 GBHO01007559 JAG36045.1 CM000361 CM002910 EDX03262.1 KMY87368.1 GEBQ01009152 JAT30825.1 CH954177 EDV57272.1 GEBQ01024836 JAT15141.1 CH480829 EDW45441.1 CH902618 EDV41270.1 AE014134 AY058512 AAF51458.1 AAL13741.1 OUUW01000004 SPP79136.1 CM000157 EDW87181.1 GFDL01014603 JAV20442.1 GAKP01007517 JAC51435.1 GAMC01019879 JAB86676.1 CH479180 EDW29032.1 GDHF01017374 JAI34940.1 DS233504 EDS30287.1 ADMH02001488 ETN62371.1 CVRI01000054 CRL00412.1 DS232303 EDS39489.1 CH379060 EAL33802.2 CH477305 EAT44146.1 CH963920 EDW77698.1 JXUM01110641 KQ565430 KXJ70985.1 GDIQ01020422 JAN74315.1 GDIP01175534 JAJ47868.1 GDIP01134971 LRGB01000243 JAL68743.1 KZS20218.1 GDIP01136224 JAL67490.1 CH916368 EDW03451.1 PNF19193.1 GL732749 EFX65346.1 CP012523 ALC38451.1 ATLV01004128 KE524190 KFB34996.1 CH940649 EDW65024.1 GGMR01007001 MBY19620.1 AJVK01017752 AJVK01017753 GFXV01003476 MBW15281.1 GFDF01004978 JAV09106.1 AXCM01004774 AJWK01001311 ABLF02033919 AXCN02000180 APCN01002131 JXJN01023846 CH933807 EDW13063.1 GECL01000728 JAP05396.1 CCAG010011565 EZ423282 ADD19558.1 JRES01000753 KNC28691.1 ADTU01003923 ADTU01003924 NNAY01000250 OXU29706.1 LJIG01022779 KRT78758.1 GDIP01061448 JAM42267.1 KQ976701 KYM77318.1 KQ977220 KYN04865.1 AAAB01005643 EAA02581.4 GGMS01001858 MBY71061.1 GL766050 EFZ15649.1 KQ981768 KYN35917.1 KQ982843 KYQ50034.1 KK107151 EZA57309.1 GDIP01144280 JAJ79122.1 GL888608 EGI59148.1 KQ980228 KYN17151.1 KQ434864 KZC09088.1 LNIX01000002 OXA60856.1 GDIQ01036372 JAN58365.1
SOQ34447.1 RSAL01000028 RVE51878.1 KQ459603 KPI92959.1 KQ461194 KPJ06880.1 AGBW02014975 OWR40789.1 JTDY01002689 KOB70916.1 GAIX01004561 JAA87999.1 KK852424 KDR24181.1 NEVH01021938 PNF19192.1 GEDC01000520 JAS36778.1 GECU01010985 JAS96721.1 ACPB03004178 GFTR01006818 JAW09608.1 QCYY01001184 ROT79860.1 GBHO01007559 JAG36045.1 CM000361 CM002910 EDX03262.1 KMY87368.1 GEBQ01009152 JAT30825.1 CH954177 EDV57272.1 GEBQ01024836 JAT15141.1 CH480829 EDW45441.1 CH902618 EDV41270.1 AE014134 AY058512 AAF51458.1 AAL13741.1 OUUW01000004 SPP79136.1 CM000157 EDW87181.1 GFDL01014603 JAV20442.1 GAKP01007517 JAC51435.1 GAMC01019879 JAB86676.1 CH479180 EDW29032.1 GDHF01017374 JAI34940.1 DS233504 EDS30287.1 ADMH02001488 ETN62371.1 CVRI01000054 CRL00412.1 DS232303 EDS39489.1 CH379060 EAL33802.2 CH477305 EAT44146.1 CH963920 EDW77698.1 JXUM01110641 KQ565430 KXJ70985.1 GDIQ01020422 JAN74315.1 GDIP01175534 JAJ47868.1 GDIP01134971 LRGB01000243 JAL68743.1 KZS20218.1 GDIP01136224 JAL67490.1 CH916368 EDW03451.1 PNF19193.1 GL732749 EFX65346.1 CP012523 ALC38451.1 ATLV01004128 KE524190 KFB34996.1 CH940649 EDW65024.1 GGMR01007001 MBY19620.1 AJVK01017752 AJVK01017753 GFXV01003476 MBW15281.1 GFDF01004978 JAV09106.1 AXCM01004774 AJWK01001311 ABLF02033919 AXCN02000180 APCN01002131 JXJN01023846 CH933807 EDW13063.1 GECL01000728 JAP05396.1 CCAG010011565 EZ423282 ADD19558.1 JRES01000753 KNC28691.1 ADTU01003923 ADTU01003924 NNAY01000250 OXU29706.1 LJIG01022779 KRT78758.1 GDIP01061448 JAM42267.1 KQ976701 KYM77318.1 KQ977220 KYN04865.1 AAAB01005643 EAA02581.4 GGMS01001858 MBY71061.1 GL766050 EFZ15649.1 KQ981768 KYN35917.1 KQ982843 KYQ50034.1 KK107151 EZA57309.1 GDIP01144280 JAJ79122.1 GL888608 EGI59148.1 KQ980228 KYN17151.1 KQ434864 KZC09088.1 LNIX01000002 OXA60856.1 GDIQ01036372 JAN58365.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000027135 UP000235965 UP000015103 UP000192221 UP000283509 UP000000304 UP000008711 UP000001292 UP000007801 UP000000803 UP000268350 UP000002282 UP000008744 UP000002320 UP000000673 UP000183832 UP000095300 UP000001819 UP000008820 UP000007798 UP000069940 UP000249989 UP000076858 UP000001070 UP000000305 UP000092553 UP000030765 UP000008792 UP000075885 UP000076408 UP000095301 UP000075901 UP000092462 UP000075920 UP000075883 UP000092461 UP000007819 UP000075886 UP000075840 UP000092460 UP000075882 UP000076407 UP000075902 UP000075903 UP000009192 UP000092444 UP000037069 UP000092445 UP000005205 UP000215335 UP000002358 UP000092443 UP000075884 UP000078540 UP000078542 UP000078200 UP000007062 UP000078541 UP000075809 UP000069272 UP000053097 UP000075900 UP000007755 UP000078492 UP000076502 UP000198287
UP000037510 UP000027135 UP000235965 UP000015103 UP000192221 UP000283509 UP000000304 UP000008711 UP000001292 UP000007801 UP000000803 UP000268350 UP000002282 UP000008744 UP000002320 UP000000673 UP000183832 UP000095300 UP000001819 UP000008820 UP000007798 UP000069940 UP000249989 UP000076858 UP000001070 UP000000305 UP000092553 UP000030765 UP000008792 UP000075885 UP000076408 UP000095301 UP000075901 UP000092462 UP000075920 UP000075883 UP000092461 UP000007819 UP000075886 UP000075840 UP000092460 UP000075882 UP000076407 UP000075902 UP000075903 UP000009192 UP000092444 UP000037069 UP000092445 UP000005205 UP000215335 UP000002358 UP000092443 UP000075884 UP000078540 UP000078542 UP000078200 UP000007062 UP000078541 UP000075809 UP000069272 UP000053097 UP000075900 UP000007755 UP000078492 UP000076502 UP000198287
PRIDE
Pfam
Interpro
IPR011545
DEAD/DEAH_box_helicase_dom
+ More
IPR014014 RNA_helicase_DEAD_Q_motif
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR027417 P-loop_NTPase
IPR032135 DUF4817
IPR037213 Run_dom_sf
IPR035969 Rab-GTPase_TBC_sf
IPR004012 Run_dom
IPR037745 SGSM1/2
IPR021935 SGSM1/2_RBD
IPR000195 Rab-GTPase-TBC_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR027417 P-loop_NTPase
IPR032135 DUF4817
IPR037213 Run_dom_sf
IPR035969 Rab-GTPase_TBC_sf
IPR004012 Run_dom
IPR037745 SGSM1/2
IPR021935 SGSM1/2_RBD
IPR000195 Rab-GTPase-TBC_dom
ProteinModelPortal
H9JLV8
A0A2W1BJ26
A0A2A4K814
A0A2H1V0T2
A0A3S2NY02
A0A194PJQ1
+ More
A0A194QPH8 A0A212EH25 A0A0L7L6I0 S4PBL6 A0A067RK13 A0A2J7PS80 A0A1B6EFR9 A0A1B6JBV5 T1IFZ3 A0A1W4V3N9 A0A224XIG1 A0A3R7QIC6 A0A0A9Z2K7 B4Q6J6 A0A1B6M4H1 B3N885 A0A1B6KUR4 B4ID15 B3MAI3 Q9VPT3 A0A3B0K0J5 B4P2X4 A0A1Q3EYR4 A0A034WBB6 W8B0D8 B4G9K2 A0A0K8V8M1 B0XJF6 W5JE88 A0A1J1ILH0 B0X2Y7 A0A1I8Q410 Q29M92 Q17CQ0 B4N0T5 A0A182H4Z1 A0A0P6HED8 A0A0P5CH29 A0A0P5T140 A0A0P5SYF6 B4JDT1 A0A2J7PS79 E9HSC9 A0A0M4E8R6 A0A084VAK2 B4LTQ1 A0A182PT25 A0A182YHI1 A0A1I8MM25 A0A2S2NQZ6 A0A182SY94 A0A1B0GQP0 A0A2H8TMY3 A0A1L8DRL1 A0A182VWM6 A0A182LVC9 A0A1B0C8R6 J9JJJ7 A0A182Q8K6 A0A182HJ83 A0A1B0C103 A0A182LCF1 A0A182XE40 A0A182TYQ9 A0A182UPG5 B4KF80 A0A0V0GBP4 D3TPD0 A0A0L0C8U0 A0A1B0A7M7 A0A158NZ67 A0A232FG16 K7IQP8 A0A1A9YIR9 A0A0T6AUP4 A0A0P5YDA7 A0A182N510 A0A195AYK6 A0A195CW58 A0A1A9UNS1 Q7QM14 A0A2S2Q052 E9IUV1 A0A195F6U8 A0A151WQ97 A0A182FKU6 A0A026WQB9 A0A0P5EDW2 A0A182RC47 F4X348 A0A151J3Z5 A0A154PCU6 A0A226EW59 A0A0N8EDW2
A0A194QPH8 A0A212EH25 A0A0L7L6I0 S4PBL6 A0A067RK13 A0A2J7PS80 A0A1B6EFR9 A0A1B6JBV5 T1IFZ3 A0A1W4V3N9 A0A224XIG1 A0A3R7QIC6 A0A0A9Z2K7 B4Q6J6 A0A1B6M4H1 B3N885 A0A1B6KUR4 B4ID15 B3MAI3 Q9VPT3 A0A3B0K0J5 B4P2X4 A0A1Q3EYR4 A0A034WBB6 W8B0D8 B4G9K2 A0A0K8V8M1 B0XJF6 W5JE88 A0A1J1ILH0 B0X2Y7 A0A1I8Q410 Q29M92 Q17CQ0 B4N0T5 A0A182H4Z1 A0A0P6HED8 A0A0P5CH29 A0A0P5T140 A0A0P5SYF6 B4JDT1 A0A2J7PS79 E9HSC9 A0A0M4E8R6 A0A084VAK2 B4LTQ1 A0A182PT25 A0A182YHI1 A0A1I8MM25 A0A2S2NQZ6 A0A182SY94 A0A1B0GQP0 A0A2H8TMY3 A0A1L8DRL1 A0A182VWM6 A0A182LVC9 A0A1B0C8R6 J9JJJ7 A0A182Q8K6 A0A182HJ83 A0A1B0C103 A0A182LCF1 A0A182XE40 A0A182TYQ9 A0A182UPG5 B4KF80 A0A0V0GBP4 D3TPD0 A0A0L0C8U0 A0A1B0A7M7 A0A158NZ67 A0A232FG16 K7IQP8 A0A1A9YIR9 A0A0T6AUP4 A0A0P5YDA7 A0A182N510 A0A195AYK6 A0A195CW58 A0A1A9UNS1 Q7QM14 A0A2S2Q052 E9IUV1 A0A195F6U8 A0A151WQ97 A0A182FKU6 A0A026WQB9 A0A0P5EDW2 A0A182RC47 F4X348 A0A151J3Z5 A0A154PCU6 A0A226EW59 A0A0N8EDW2
PDB
5IVL
E-value=1.90481e-38,
Score=401
Ontologies
GO
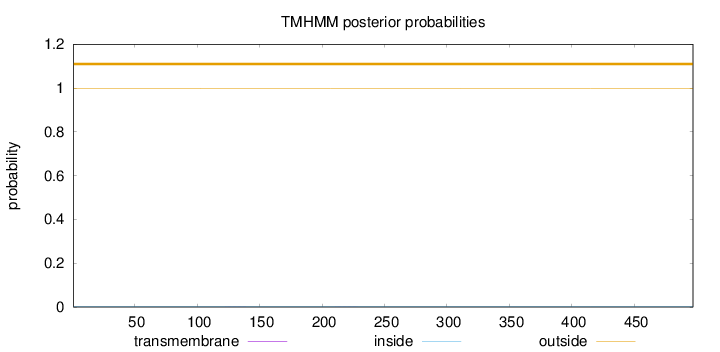
Topology
Length:
497
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0083
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00344
outside
1 - 497
Population Genetic Test Statistics
Pi
24.221652
Theta
21.203329
Tajima's D
-0.963877
CLR
0.958607
CSRT
0.142892855357232
Interpretation
Uncertain
