Gene
KWMTBOMO07024
Pre Gene Modal
BGIBMGA010462
Annotation
PREDICTED:_acylglycerol_kinase?_mitochondrial_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.346
Sequence
CDS
ATGGTACATATATCATCTGTTTCAGGATTGTTAAGACGAAATGATCAAGCAAACATGTTTCCAGTAGGAGTGTTGCCACTCGGACGCACAAACTCAATCGGTAACACTTTATTTCCAAACGGTAAGGGTGTACAAAAAGTTAAGCAGTTGATCACCGCCTCAATGGCAATCATCAAAGGAAACACAGTATGGAAAGATGCCATGAAAATAGAACAAGTTACTGATGAGTCTGAAACTCCTAATCGTCCGATTTACGCTATGGCATCAATGGAGTGGGGAGCTTTTAGGGACACCCTAGCAAAGAAGGATAAGTACTGGTTCTATGGTCCTTTAAGAGAGTATGCTGCCTACATCTTCAATGGTTACAAGAGTTCATTAAACTGGAATTGCAGCGGCACCATCAAATATACTCCTCCGTGTGTCGGGTGCACCAACTGTACAGCTAAAAGGCCGGAAGTAAAACGAAAATGGTTCTTTGTTTCGTCCACCGAAGCGGCTGGGCAGCTTGACCAAACGAAGATTCTAAATCCAGAGTGCCTGACATCGAAAGAACTGTGCTTTCGAACTTGCGATTTCAAAATTAAAACGCAGAATGTTGCAATGAACCATCAGATACCAGCCTTGAGTATATTTCTGGGCAGAAACAGTTACTCATACACCGACTTTGTGTCTGAAGGTTGGCGCAGAATAAAGACAAACTCGGATCCAGAGCCAATTTTTGCACGTACCATCGAGATCACACCTAAAGAAACGGATAGAGATACAGTTATAGAAATCGACAAGGAGGATTTTGATATGAAACCAATTAAAATAACGTTAATGCCAAAAGTTATTAAATTATTCTGTCAATAA
Protein
MVHISSVSGLLRRNDQANMFPVGVLPLGRTNSIGNTLFPNGKGVQKVKQLITASMAIIKGNTVWKDAMKIEQVTDESETPNRPIYAMASMEWGAFRDTLAKKDKYWFYGPLREYAAYIFNGYKSSLNWNCSGTIKYTPPCVGCTNCTAKRPEVKRKWFFVSSTEAAGQLDQTKILNPECLTSKELCFRTCDFKIKTQNVAMNHQIPALSIFLGRNSYSYTDFVSEGWRRIKTNSDPEPIFARTIEITPKETDRDTVIEIDKEDFDMKPIKITLMPKVIKLFCQ
Summary
Uniprot
H9JLR0
A0A0L7LUV8
A0A3S2NMG5
A0A194QMX8
A0A194PI17
A0A2A4K801
+ More
A0A212EIG8 A0A2H1V0S5 A0A2W1BFV5 A0A2H1W2F6 A0A1Y1MH29 A0A1W4XH92 J3JXF9 U4UNA3 K7J048 A0A232FHW7 A0A158NRB4 A0A0L7RH66 E9IS08 A0A195FIS5 A0A151XHM6 E2AUW3 A0A0J7KI54 F4WAZ7 A0A2A3EBT2 A0A088ANE4 E2C529 A0A026X3X2 A0A3L8DJ65 A0A1B0D3G0 A0A154PD31 A0A182FGV1 A0A2M4BNU5 A0A2M4BMY7 A0A1L8DYU9 A0A2M4BMZ1 A0A1B0CE40 A0A1L8DZ26 A0A182ITG8 A0A2M4ANB0 A0A182WL88 A0A182YJ99 W5JWA5 A0A2J7PMX4 A0A2J7PMV4 A0A182NJG7 A0A182R076 A0A182I286 A0A182V9E7 A0A182UI69 A0A182KQ16 A0A182H022 A0A182XMF6 A0A182PIF3 A0A182RFH6 B0W3S0 Q7Q6I5 A0A1Q3F6T5 T1GI02 A0A1S4FKF5 E9GYN2 A0A1B6E9W4 A0A0P5A4H4 Q16Y30 A0A023ES09 A0A0P5A726 A0A0P5PLL0 A0A0P5FJR4 A0A1J1IAT1 E0VN09 A0A2S2QPA9 A0A084WDC4 A0A0M8ZRQ8 A0A336LLJ1 A0A2S2P101 A0A195BFE7 J9K012 A0A0P6IAA4 A0A0P6BJL7 A0A0P5Z4G6 A0A2H8TEM6 A0A2P8YCT2 A0A0P6CQ75 A0A146KZL6 A0A182MPD4 A0A0A9Y733 A0A146L1A1 A0A1B6FJP8 A0A1B6F5Z7 A0A067R3G9 A0A0N7ZC87 A0A0P5DSU5 A0A224XN73 A0A023F7X3 A0A0V0GAU7 B4KHN8 A0A0P5QMD4 A0A0P5MXI9 A0A0P5TEX1
A0A212EIG8 A0A2H1V0S5 A0A2W1BFV5 A0A2H1W2F6 A0A1Y1MH29 A0A1W4XH92 J3JXF9 U4UNA3 K7J048 A0A232FHW7 A0A158NRB4 A0A0L7RH66 E9IS08 A0A195FIS5 A0A151XHM6 E2AUW3 A0A0J7KI54 F4WAZ7 A0A2A3EBT2 A0A088ANE4 E2C529 A0A026X3X2 A0A3L8DJ65 A0A1B0D3G0 A0A154PD31 A0A182FGV1 A0A2M4BNU5 A0A2M4BMY7 A0A1L8DYU9 A0A2M4BMZ1 A0A1B0CE40 A0A1L8DZ26 A0A182ITG8 A0A2M4ANB0 A0A182WL88 A0A182YJ99 W5JWA5 A0A2J7PMX4 A0A2J7PMV4 A0A182NJG7 A0A182R076 A0A182I286 A0A182V9E7 A0A182UI69 A0A182KQ16 A0A182H022 A0A182XMF6 A0A182PIF3 A0A182RFH6 B0W3S0 Q7Q6I5 A0A1Q3F6T5 T1GI02 A0A1S4FKF5 E9GYN2 A0A1B6E9W4 A0A0P5A4H4 Q16Y30 A0A023ES09 A0A0P5A726 A0A0P5PLL0 A0A0P5FJR4 A0A1J1IAT1 E0VN09 A0A2S2QPA9 A0A084WDC4 A0A0M8ZRQ8 A0A336LLJ1 A0A2S2P101 A0A195BFE7 J9K012 A0A0P6IAA4 A0A0P6BJL7 A0A0P5Z4G6 A0A2H8TEM6 A0A2P8YCT2 A0A0P6CQ75 A0A146KZL6 A0A182MPD4 A0A0A9Y733 A0A146L1A1 A0A1B6FJP8 A0A1B6F5Z7 A0A067R3G9 A0A0N7ZC87 A0A0P5DSU5 A0A224XN73 A0A023F7X3 A0A0V0GAU7 B4KHN8 A0A0P5QMD4 A0A0P5MXI9 A0A0P5TEX1
Pubmed
19121390
26227816
26354079
22118469
28756777
28004739
+ More
22516182 23537049 20075255 28648823 21347285 21282665 20798317 21719571 24508170 30249741 25244985 20920257 23761445 20966253 26483478 12364791 21292972 17510324 24945155 20566863 24438588 29403074 26823975 25401762 24845553 25474469 17994087
22516182 23537049 20075255 28648823 21347285 21282665 20798317 21719571 24508170 30249741 25244985 20920257 23761445 20966253 26483478 12364791 21292972 17510324 24945155 20566863 24438588 29403074 26823975 25401762 24845553 25474469 17994087
EMBL
BABH01005221
JTDY01000041
KOB79225.1
RSAL01000003
RVE54723.1
KQ461194
+ More
KPJ06877.1 KQ459603 KPI92962.1 NWSH01000039 PCG80367.1 AGBW02014643 OWR41282.1 ODYU01000155 SOQ34450.1 KZ150274 PZC71660.1 ODYU01005904 SOQ47275.1 GEZM01031035 GEZM01031034 JAV85109.1 BT127928 AEE62890.1 KB632433 ERL95569.1 NNAY01000199 OXU30059.1 ADTU01023845 KQ414596 KOC70051.1 GL765283 EFZ16687.1 KQ981522 KYN40565.1 KQ982117 KYQ59903.1 GL442898 EFN62771.1 LBMM01007252 KMQ89901.1 GL888056 EGI68617.1 KZ288292 PBC29195.1 GL452737 EFN76937.1 KK107019 EZA62691.1 QOIP01000007 RLU20391.1 AJVK01023535 KQ434875 KZC09762.1 GGFJ01005307 MBW54448.1 GGFJ01005308 MBW54449.1 GFDF01002441 JAV11643.1 GGFJ01005309 MBW54450.1 AJWK01008544 GFDF01002354 JAV11730.1 GGFK01008944 MBW42265.1 ADMH02000243 ETN67259.1 NEVH01023960 PNF17669.1 PNF17671.1 AXCN02000446 APCN01000128 JXUM01100575 KQ564580 KXJ72072.1 DS231833 EDS32217.1 AAAB01008960 EAA11370.4 GFDL01011806 JAV23239.1 CAQQ02390494 GL732575 EFX75428.1 GEDC01002569 JAS34729.1 GDIP01204094 JAJ19308.1 CH477526 EAT39530.1 GAPW01001590 JAC12008.1 GDIP01204093 JAJ19309.1 GDIQ01128847 JAL22879.1 GDIP01147351 JAJ76051.1 CVRI01000042 CRK95545.1 DS235331 EEB14765.1 GGMS01010406 MBY79609.1 ATLV01022986 KE525339 KFB48218.1 KQ435944 KOX68189.1 UFQS01000007 UFQT01000007 SSW97117.1 SSX17503.1 GGMR01009987 MBY22606.1 KQ976504 KYM82912.1 ABLF02039965 ABLF02039972 GDIQ01014376 JAN80361.1 GDIP01017019 JAM86696.1 GDIP01049395 JAM54320.1 GFXV01000738 MBW12543.1 PYGN01000700 PSN42053.1 GDIQ01088649 JAN06088.1 GDHC01018197 JAQ00432.1 AXCM01004988 GBHO01016168 JAG27436.1 GDHC01016278 JAQ02351.1 GECZ01019372 JAS50397.1 GECZ01024164 JAS45605.1 KK852936 KDR13605.1 GDRN01071070 GDRN01071069 JAI63760.1 GDIP01151148 JAJ72254.1 GFTR01006394 JAW10032.1 GBBI01001374 JAC17338.1 GECL01001135 JAP04989.1 CH933807 EDW12317.2 GDIQ01132844 JAL18882.1 GDIQ01150382 JAL01344.1 GDIP01127795 JAL75919.1
KPJ06877.1 KQ459603 KPI92962.1 NWSH01000039 PCG80367.1 AGBW02014643 OWR41282.1 ODYU01000155 SOQ34450.1 KZ150274 PZC71660.1 ODYU01005904 SOQ47275.1 GEZM01031035 GEZM01031034 JAV85109.1 BT127928 AEE62890.1 KB632433 ERL95569.1 NNAY01000199 OXU30059.1 ADTU01023845 KQ414596 KOC70051.1 GL765283 EFZ16687.1 KQ981522 KYN40565.1 KQ982117 KYQ59903.1 GL442898 EFN62771.1 LBMM01007252 KMQ89901.1 GL888056 EGI68617.1 KZ288292 PBC29195.1 GL452737 EFN76937.1 KK107019 EZA62691.1 QOIP01000007 RLU20391.1 AJVK01023535 KQ434875 KZC09762.1 GGFJ01005307 MBW54448.1 GGFJ01005308 MBW54449.1 GFDF01002441 JAV11643.1 GGFJ01005309 MBW54450.1 AJWK01008544 GFDF01002354 JAV11730.1 GGFK01008944 MBW42265.1 ADMH02000243 ETN67259.1 NEVH01023960 PNF17669.1 PNF17671.1 AXCN02000446 APCN01000128 JXUM01100575 KQ564580 KXJ72072.1 DS231833 EDS32217.1 AAAB01008960 EAA11370.4 GFDL01011806 JAV23239.1 CAQQ02390494 GL732575 EFX75428.1 GEDC01002569 JAS34729.1 GDIP01204094 JAJ19308.1 CH477526 EAT39530.1 GAPW01001590 JAC12008.1 GDIP01204093 JAJ19309.1 GDIQ01128847 JAL22879.1 GDIP01147351 JAJ76051.1 CVRI01000042 CRK95545.1 DS235331 EEB14765.1 GGMS01010406 MBY79609.1 ATLV01022986 KE525339 KFB48218.1 KQ435944 KOX68189.1 UFQS01000007 UFQT01000007 SSW97117.1 SSX17503.1 GGMR01009987 MBY22606.1 KQ976504 KYM82912.1 ABLF02039965 ABLF02039972 GDIQ01014376 JAN80361.1 GDIP01017019 JAM86696.1 GDIP01049395 JAM54320.1 GFXV01000738 MBW12543.1 PYGN01000700 PSN42053.1 GDIQ01088649 JAN06088.1 GDHC01018197 JAQ00432.1 AXCM01004988 GBHO01016168 JAG27436.1 GDHC01016278 JAQ02351.1 GECZ01019372 JAS50397.1 GECZ01024164 JAS45605.1 KK852936 KDR13605.1 GDRN01071070 GDRN01071069 JAI63760.1 GDIP01151148 JAJ72254.1 GFTR01006394 JAW10032.1 GBBI01001374 JAC17338.1 GECL01001135 JAP04989.1 CH933807 EDW12317.2 GDIQ01132844 JAL18882.1 GDIQ01150382 JAL01344.1 GDIP01127795 JAL75919.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000053240
UP000053268
UP000218220
+ More
UP000007151 UP000192223 UP000030742 UP000002358 UP000215335 UP000005205 UP000053825 UP000078541 UP000075809 UP000000311 UP000036403 UP000007755 UP000242457 UP000005203 UP000008237 UP000053097 UP000279307 UP000092462 UP000076502 UP000069272 UP000092461 UP000075880 UP000075920 UP000076408 UP000000673 UP000235965 UP000075884 UP000075886 UP000075840 UP000075903 UP000075902 UP000075882 UP000069940 UP000249989 UP000076407 UP000075885 UP000075900 UP000002320 UP000007062 UP000015102 UP000000305 UP000008820 UP000183832 UP000009046 UP000030765 UP000053105 UP000078540 UP000007819 UP000245037 UP000075883 UP000027135 UP000009192
UP000007151 UP000192223 UP000030742 UP000002358 UP000215335 UP000005205 UP000053825 UP000078541 UP000075809 UP000000311 UP000036403 UP000007755 UP000242457 UP000005203 UP000008237 UP000053097 UP000279307 UP000092462 UP000076502 UP000069272 UP000092461 UP000075880 UP000075920 UP000076408 UP000000673 UP000235965 UP000075884 UP000075886 UP000075840 UP000075903 UP000075902 UP000075882 UP000069940 UP000249989 UP000076407 UP000075885 UP000075900 UP000002320 UP000007062 UP000015102 UP000000305 UP000008820 UP000183832 UP000009046 UP000030765 UP000053105 UP000078540 UP000007819 UP000245037 UP000075883 UP000027135 UP000009192
PRIDE
Interpro
SUPFAM
SSF111331
SSF111331
Gene 3D
CDD
ProteinModelPortal
H9JLR0
A0A0L7LUV8
A0A3S2NMG5
A0A194QMX8
A0A194PI17
A0A2A4K801
+ More
A0A212EIG8 A0A2H1V0S5 A0A2W1BFV5 A0A2H1W2F6 A0A1Y1MH29 A0A1W4XH92 J3JXF9 U4UNA3 K7J048 A0A232FHW7 A0A158NRB4 A0A0L7RH66 E9IS08 A0A195FIS5 A0A151XHM6 E2AUW3 A0A0J7KI54 F4WAZ7 A0A2A3EBT2 A0A088ANE4 E2C529 A0A026X3X2 A0A3L8DJ65 A0A1B0D3G0 A0A154PD31 A0A182FGV1 A0A2M4BNU5 A0A2M4BMY7 A0A1L8DYU9 A0A2M4BMZ1 A0A1B0CE40 A0A1L8DZ26 A0A182ITG8 A0A2M4ANB0 A0A182WL88 A0A182YJ99 W5JWA5 A0A2J7PMX4 A0A2J7PMV4 A0A182NJG7 A0A182R076 A0A182I286 A0A182V9E7 A0A182UI69 A0A182KQ16 A0A182H022 A0A182XMF6 A0A182PIF3 A0A182RFH6 B0W3S0 Q7Q6I5 A0A1Q3F6T5 T1GI02 A0A1S4FKF5 E9GYN2 A0A1B6E9W4 A0A0P5A4H4 Q16Y30 A0A023ES09 A0A0P5A726 A0A0P5PLL0 A0A0P5FJR4 A0A1J1IAT1 E0VN09 A0A2S2QPA9 A0A084WDC4 A0A0M8ZRQ8 A0A336LLJ1 A0A2S2P101 A0A195BFE7 J9K012 A0A0P6IAA4 A0A0P6BJL7 A0A0P5Z4G6 A0A2H8TEM6 A0A2P8YCT2 A0A0P6CQ75 A0A146KZL6 A0A182MPD4 A0A0A9Y733 A0A146L1A1 A0A1B6FJP8 A0A1B6F5Z7 A0A067R3G9 A0A0N7ZC87 A0A0P5DSU5 A0A224XN73 A0A023F7X3 A0A0V0GAU7 B4KHN8 A0A0P5QMD4 A0A0P5MXI9 A0A0P5TEX1
A0A212EIG8 A0A2H1V0S5 A0A2W1BFV5 A0A2H1W2F6 A0A1Y1MH29 A0A1W4XH92 J3JXF9 U4UNA3 K7J048 A0A232FHW7 A0A158NRB4 A0A0L7RH66 E9IS08 A0A195FIS5 A0A151XHM6 E2AUW3 A0A0J7KI54 F4WAZ7 A0A2A3EBT2 A0A088ANE4 E2C529 A0A026X3X2 A0A3L8DJ65 A0A1B0D3G0 A0A154PD31 A0A182FGV1 A0A2M4BNU5 A0A2M4BMY7 A0A1L8DYU9 A0A2M4BMZ1 A0A1B0CE40 A0A1L8DZ26 A0A182ITG8 A0A2M4ANB0 A0A182WL88 A0A182YJ99 W5JWA5 A0A2J7PMX4 A0A2J7PMV4 A0A182NJG7 A0A182R076 A0A182I286 A0A182V9E7 A0A182UI69 A0A182KQ16 A0A182H022 A0A182XMF6 A0A182PIF3 A0A182RFH6 B0W3S0 Q7Q6I5 A0A1Q3F6T5 T1GI02 A0A1S4FKF5 E9GYN2 A0A1B6E9W4 A0A0P5A4H4 Q16Y30 A0A023ES09 A0A0P5A726 A0A0P5PLL0 A0A0P5FJR4 A0A1J1IAT1 E0VN09 A0A2S2QPA9 A0A084WDC4 A0A0M8ZRQ8 A0A336LLJ1 A0A2S2P101 A0A195BFE7 J9K012 A0A0P6IAA4 A0A0P6BJL7 A0A0P5Z4G6 A0A2H8TEM6 A0A2P8YCT2 A0A0P6CQ75 A0A146KZL6 A0A182MPD4 A0A0A9Y733 A0A146L1A1 A0A1B6FJP8 A0A1B6F5Z7 A0A067R3G9 A0A0N7ZC87 A0A0P5DSU5 A0A224XN73 A0A023F7X3 A0A0V0GAU7 B4KHN8 A0A0P5QMD4 A0A0P5MXI9 A0A0P5TEX1
Ontologies
KEGG
PANTHER
Topology
Subcellular location
Nucleus
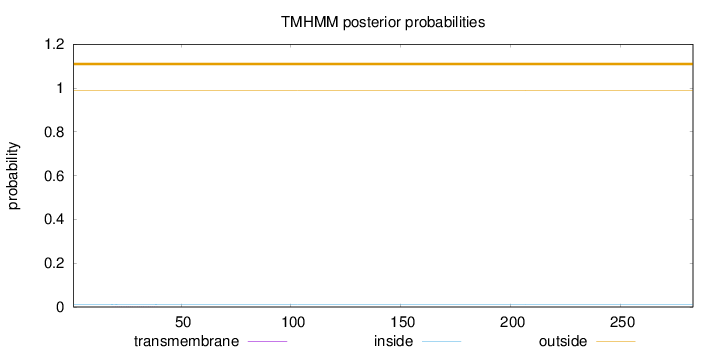
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01255
Exp number, first 60 AAs:
0.01026
Total prob of N-in:
0.01114
outside
1 - 283
Population Genetic Test Statistics
Pi
20.741327
Theta
19.622717
Tajima's D
0.173274
CLR
0.349272
CSRT
0.425628718564072
Interpretation
Uncertain
