Gene
KWMTBOMO07023
Pre Gene Modal
BGIBMGA010508
Annotation
PREDICTED:_probable_methyltransferase_TARBP1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.886
Sequence
CDS
ATGTACCGAATTGATCCACGAATAAAGAAATCTTTAGCCGCGAATATAGGAATTGATTCGCATTTTTTTAAATCTGCGAATTCTGAGAAAGCCAATGAAGGAAGTGCGAATAAGAAAGAGATCATCGGAGACTTGATAGTGGTAGCTTCTTTAGTAGATAAATTGCCGAACTTGGGCGGCATGGCCCGGACCAGCGAGGTGTTCGGGGTGAGGAGCTACGTGGTGGGCAGTCTGCGGCACCTCCAGGACAAGCAATTCCAGGGATTGAGTGTGTCAGCCGAGCGATGGATAGACGTTGAGGAAGTGAGACCCGGTCAGCCTCTGAAAGAGTTTCTGGCCAACAAGAAGTCGGAAGGTTACTCTGTGGTGGCAGCCGAGCAAACGTCGACCAGCGTCAAACTGGAAAGCTTTAAATTTCCAAAGAAAACTGTTTTATTACTCGGACACGAGAAGGAAGGCGTGCCGTGCGACCTGCTGCCGCTCATGGACCAGTGCGTGGAGATCCCGCAGCGCGGCGTCGTGCGCTCGCTCAACGTGCACGTCGCCGCCGCCATATTCGTCTGGGAGTACACCAGGCAAAACGTCTTGTGA
Protein
MYRIDPRIKKSLAANIGIDSHFFKSANSEKANEGSANKKEIIGDLIVVASLVDKLPNLGGMARTSEVFGVRSYVVGSLRHLQDKQFQGLSVSAERWIDVEEVRPGQPLKEFLANKKSEGYSVVAAEQTSTSVKLESFKFPKKTVLLLGHEKEGVPCDLLPLMDQCVEIPQRGVVRSLNVHVAAAIFVWEYTRQNVL
Summary
Uniprot
H9JLV6
A0A0L7LUU5
A0A2H1W2V7
A0A2A4K8N7
A0A194QTK3
A0A194PIT7
+ More
A0A2J7R4Z6 V5HMX9 A0A2J7R4Y4 A0A2J7R4Y6 A0A2R5L604 L7MGU6 L7MGL0 E9H6E6 A0A0S7HMT4 A0A147BM09 A0A2J7R4Y8 B8A4Z4 A0A147BCS0 A0A0S7HQV1 A0A131YUS7 A0A1B6IZ52 A0A067RAR9 A7RT83 A0A2D4KT68 A0A087V057 A0A1A7XUZ7 E2A8V6 A0A0P5IQV3 A0A1A8PN31 A0A0P4ZKG1 A0A0P5VNE1 A0A0P5SW57 A0A0P4X8I4 A0A0N8CNW5 A0A0P6C8V0 A0A2C9JQH1 A0A0P5D1K9 A0A3B4B7G3 A0A151M2V4 Q0WLH7 A0A0P6HCJ9 A0A0P5N1P9 A0A0P6CZY3 A0A1A8M905 A0A3P9QIT3 A0A315VS85 F1Q8L2 A0A3B3TT16 A0A224XMH8 A0A3B3XK19 A0A3B3T950 K9KA80 A0A154NWE1 K1P8Q1 A0A2G8K9L9 A0A1A8HCX1 A0A3Q1C828 A0A3P9BC93 A0A3B4GW00 H2M436 S7NKP6 A0A3P9MKE3 I3JTV2 A0A3Q3BVS6 A0A1Y1MDV7 A0A3P9ITH8 L5K0I2 A0A1U8DDP0 M3XUM3 A0A1Q3DI49 A0A3P8V522 A0A1Q3DIS9 A0A3P8UYI1 A0A3Q0HAU2 A0A1A8IGW6 A0A3Q0HF71 A0A1A7WIE5 A0A1A7YB09 A0A1U8DGC2 A0A151Z8P4 A0A3B5K2I8 A0A1U8DNQ3 A0A3B3B6H9 A0A3P8NDU6 A0A3Q3F6X1 A0A1S3F1D8 A0A3Q0SW69 A0A226P673 A0A384C0B7 G1L9G6 D2I143 H0ZIX2 A0A1A8KAV7 A0A3Q3SXK3 A0A091JKD9 A0A3Q1JFH6 A0A3Q1FCJ3
A0A2J7R4Z6 V5HMX9 A0A2J7R4Y4 A0A2J7R4Y6 A0A2R5L604 L7MGU6 L7MGL0 E9H6E6 A0A0S7HMT4 A0A147BM09 A0A2J7R4Y8 B8A4Z4 A0A147BCS0 A0A0S7HQV1 A0A131YUS7 A0A1B6IZ52 A0A067RAR9 A7RT83 A0A2D4KT68 A0A087V057 A0A1A7XUZ7 E2A8V6 A0A0P5IQV3 A0A1A8PN31 A0A0P4ZKG1 A0A0P5VNE1 A0A0P5SW57 A0A0P4X8I4 A0A0N8CNW5 A0A0P6C8V0 A0A2C9JQH1 A0A0P5D1K9 A0A3B4B7G3 A0A151M2V4 Q0WLH7 A0A0P6HCJ9 A0A0P5N1P9 A0A0P6CZY3 A0A1A8M905 A0A3P9QIT3 A0A315VS85 F1Q8L2 A0A3B3TT16 A0A224XMH8 A0A3B3XK19 A0A3B3T950 K9KA80 A0A154NWE1 K1P8Q1 A0A2G8K9L9 A0A1A8HCX1 A0A3Q1C828 A0A3P9BC93 A0A3B4GW00 H2M436 S7NKP6 A0A3P9MKE3 I3JTV2 A0A3Q3BVS6 A0A1Y1MDV7 A0A3P9ITH8 L5K0I2 A0A1U8DDP0 M3XUM3 A0A1Q3DI49 A0A3P8V522 A0A1Q3DIS9 A0A3P8UYI1 A0A3Q0HAU2 A0A1A8IGW6 A0A3Q0HF71 A0A1A7WIE5 A0A1A7YB09 A0A1U8DGC2 A0A151Z8P4 A0A3B5K2I8 A0A1U8DNQ3 A0A3B3B6H9 A0A3P8NDU6 A0A3Q3F6X1 A0A1S3F1D8 A0A3Q0SW69 A0A226P673 A0A384C0B7 G1L9G6 D2I143 H0ZIX2 A0A1A8KAV7 A0A3Q3SXK3 A0A091JKD9 A0A3Q1JFH6 A0A3Q1FCJ3
Pubmed
EMBL
BABH01005217
BABH01005218
BABH01005219
JTDY01000041
KOB79222.1
ODYU01005904
+ More
SOQ47276.1 NWSH01000039 PCG80366.1 KQ461194 KPJ06876.1 KQ459603 KPI92963.1 NEVH01007399 PNF35899.1 GANP01005279 JAB79189.1 PNF35894.1 PNF35896.1 GGLE01000783 MBY04909.1 GACK01002625 JAA62409.1 GACK01002626 JAA62408.1 GL732597 EFX72710.1 GBYX01439776 JAO41582.1 GEGO01003574 JAR91830.1 PNF35897.1 BX324188 GEGO01006850 JAR88554.1 GBYX01439777 JAO41581.1 GEDV01006265 JAP82292.1 GECU01015514 JAS92192.1 KK852777 KDR16784.1 DS469536 EDO45339.1 IACL01089992 LAB11899.1 KK122567 KFM82996.1 HADW01020496 SBP21896.1 GL437663 EFN70185.1 GDIQ01209253 JAK42472.1 HAEH01007646 SBR82422.1 GDIP01213218 JAJ10184.1 GDIP01097680 JAM06035.1 GDIP01134469 JAL69245.1 GDIP01247863 JAI75538.1 GDIP01113327 JAL90387.1 GDIP01007072 LRGB01003375 JAM96643.1 KZS02969.1 GDIP01164163 JAJ59239.1 AKHW03006780 KYO18816.1 AK230224 BAF02030.1 GDIQ01021163 JAN73574.1 GDIQ01148654 JAL03072.1 GDIQ01085025 JAN09712.1 HAEF01011672 SBR52789.1 NHOQ01001229 PWA26081.1 GFTR01002729 JAW13697.1 JL619649 AEP99192.1 KQ434772 KZC03901.1 JH817871 EKC20062.1 MRZV01000761 PIK44701.1 HAEC01013000 SBQ81217.1 KE164339 EPQ17140.1 AERX01006398 GEZM01034249 JAV83853.1 KB031047 ELK05090.1 AEYP01092078 BDDD01008499 GAV91983.1 BDDD01009153 GAV92223.1 HAED01009453 SBQ95665.1 HADW01004105 SBP05505.1 HADX01004898 SBP27130.1 LODT01000037 KYQ90343.1 AWGT02000162 OXB74987.1 ACTA01123540 ACTA01131540 ACTA01139540 ACTA01147539 GL193949 EFB17917.1 ABQF01014617 HAEE01008749 SBR28799.1 KK502085 KFP20373.1
SOQ47276.1 NWSH01000039 PCG80366.1 KQ461194 KPJ06876.1 KQ459603 KPI92963.1 NEVH01007399 PNF35899.1 GANP01005279 JAB79189.1 PNF35894.1 PNF35896.1 GGLE01000783 MBY04909.1 GACK01002625 JAA62409.1 GACK01002626 JAA62408.1 GL732597 EFX72710.1 GBYX01439776 JAO41582.1 GEGO01003574 JAR91830.1 PNF35897.1 BX324188 GEGO01006850 JAR88554.1 GBYX01439777 JAO41581.1 GEDV01006265 JAP82292.1 GECU01015514 JAS92192.1 KK852777 KDR16784.1 DS469536 EDO45339.1 IACL01089992 LAB11899.1 KK122567 KFM82996.1 HADW01020496 SBP21896.1 GL437663 EFN70185.1 GDIQ01209253 JAK42472.1 HAEH01007646 SBR82422.1 GDIP01213218 JAJ10184.1 GDIP01097680 JAM06035.1 GDIP01134469 JAL69245.1 GDIP01247863 JAI75538.1 GDIP01113327 JAL90387.1 GDIP01007072 LRGB01003375 JAM96643.1 KZS02969.1 GDIP01164163 JAJ59239.1 AKHW03006780 KYO18816.1 AK230224 BAF02030.1 GDIQ01021163 JAN73574.1 GDIQ01148654 JAL03072.1 GDIQ01085025 JAN09712.1 HAEF01011672 SBR52789.1 NHOQ01001229 PWA26081.1 GFTR01002729 JAW13697.1 JL619649 AEP99192.1 KQ434772 KZC03901.1 JH817871 EKC20062.1 MRZV01000761 PIK44701.1 HAEC01013000 SBQ81217.1 KE164339 EPQ17140.1 AERX01006398 GEZM01034249 JAV83853.1 KB031047 ELK05090.1 AEYP01092078 BDDD01008499 GAV91983.1 BDDD01009153 GAV92223.1 HAED01009453 SBQ95665.1 HADW01004105 SBP05505.1 HADX01004898 SBP27130.1 LODT01000037 KYQ90343.1 AWGT02000162 OXB74987.1 ACTA01123540 ACTA01131540 ACTA01139540 ACTA01147539 GL193949 EFB17917.1 ABQF01014617 HAEE01008749 SBR28799.1 KK502085 KFP20373.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000235965
+ More
UP000000305 UP000000437 UP000027135 UP000001593 UP000054359 UP000000311 UP000076858 UP000076420 UP000261520 UP000050525 UP000242638 UP000261500 UP000261480 UP000261540 UP000076502 UP000005408 UP000230750 UP000257160 UP000265160 UP000261460 UP000001038 UP000265180 UP000005207 UP000264840 UP000265200 UP000010552 UP000189705 UP000000715 UP000187406 UP000265120 UP000076078 UP000005226 UP000261560 UP000265100 UP000261660 UP000081671 UP000261340 UP000198419 UP000261680 UP000008912 UP000007754 UP000261640 UP000053119 UP000265040 UP000257200
UP000000305 UP000000437 UP000027135 UP000001593 UP000054359 UP000000311 UP000076858 UP000076420 UP000261520 UP000050525 UP000242638 UP000261500 UP000261480 UP000261540 UP000076502 UP000005408 UP000230750 UP000257160 UP000265160 UP000261460 UP000001038 UP000265180 UP000005207 UP000264840 UP000265200 UP000010552 UP000189705 UP000000715 UP000187406 UP000265120 UP000076078 UP000005226 UP000261560 UP000265100 UP000261660 UP000081671 UP000261340 UP000198419 UP000261680 UP000008912 UP000007754 UP000261640 UP000053119 UP000265040 UP000257200
Pfam
PF00588 SpoU_methylase
Interpro
Gene 3D
ProteinModelPortal
H9JLV6
A0A0L7LUU5
A0A2H1W2V7
A0A2A4K8N7
A0A194QTK3
A0A194PIT7
+ More
A0A2J7R4Z6 V5HMX9 A0A2J7R4Y4 A0A2J7R4Y6 A0A2R5L604 L7MGU6 L7MGL0 E9H6E6 A0A0S7HMT4 A0A147BM09 A0A2J7R4Y8 B8A4Z4 A0A147BCS0 A0A0S7HQV1 A0A131YUS7 A0A1B6IZ52 A0A067RAR9 A7RT83 A0A2D4KT68 A0A087V057 A0A1A7XUZ7 E2A8V6 A0A0P5IQV3 A0A1A8PN31 A0A0P4ZKG1 A0A0P5VNE1 A0A0P5SW57 A0A0P4X8I4 A0A0N8CNW5 A0A0P6C8V0 A0A2C9JQH1 A0A0P5D1K9 A0A3B4B7G3 A0A151M2V4 Q0WLH7 A0A0P6HCJ9 A0A0P5N1P9 A0A0P6CZY3 A0A1A8M905 A0A3P9QIT3 A0A315VS85 F1Q8L2 A0A3B3TT16 A0A224XMH8 A0A3B3XK19 A0A3B3T950 K9KA80 A0A154NWE1 K1P8Q1 A0A2G8K9L9 A0A1A8HCX1 A0A3Q1C828 A0A3P9BC93 A0A3B4GW00 H2M436 S7NKP6 A0A3P9MKE3 I3JTV2 A0A3Q3BVS6 A0A1Y1MDV7 A0A3P9ITH8 L5K0I2 A0A1U8DDP0 M3XUM3 A0A1Q3DI49 A0A3P8V522 A0A1Q3DIS9 A0A3P8UYI1 A0A3Q0HAU2 A0A1A8IGW6 A0A3Q0HF71 A0A1A7WIE5 A0A1A7YB09 A0A1U8DGC2 A0A151Z8P4 A0A3B5K2I8 A0A1U8DNQ3 A0A3B3B6H9 A0A3P8NDU6 A0A3Q3F6X1 A0A1S3F1D8 A0A3Q0SW69 A0A226P673 A0A384C0B7 G1L9G6 D2I143 H0ZIX2 A0A1A8KAV7 A0A3Q3SXK3 A0A091JKD9 A0A3Q1JFH6 A0A3Q1FCJ3
A0A2J7R4Z6 V5HMX9 A0A2J7R4Y4 A0A2J7R4Y6 A0A2R5L604 L7MGU6 L7MGL0 E9H6E6 A0A0S7HMT4 A0A147BM09 A0A2J7R4Y8 B8A4Z4 A0A147BCS0 A0A0S7HQV1 A0A131YUS7 A0A1B6IZ52 A0A067RAR9 A7RT83 A0A2D4KT68 A0A087V057 A0A1A7XUZ7 E2A8V6 A0A0P5IQV3 A0A1A8PN31 A0A0P4ZKG1 A0A0P5VNE1 A0A0P5SW57 A0A0P4X8I4 A0A0N8CNW5 A0A0P6C8V0 A0A2C9JQH1 A0A0P5D1K9 A0A3B4B7G3 A0A151M2V4 Q0WLH7 A0A0P6HCJ9 A0A0P5N1P9 A0A0P6CZY3 A0A1A8M905 A0A3P9QIT3 A0A315VS85 F1Q8L2 A0A3B3TT16 A0A224XMH8 A0A3B3XK19 A0A3B3T950 K9KA80 A0A154NWE1 K1P8Q1 A0A2G8K9L9 A0A1A8HCX1 A0A3Q1C828 A0A3P9BC93 A0A3B4GW00 H2M436 S7NKP6 A0A3P9MKE3 I3JTV2 A0A3Q3BVS6 A0A1Y1MDV7 A0A3P9ITH8 L5K0I2 A0A1U8DDP0 M3XUM3 A0A1Q3DI49 A0A3P8V522 A0A1Q3DIS9 A0A3P8UYI1 A0A3Q0HAU2 A0A1A8IGW6 A0A3Q0HF71 A0A1A7WIE5 A0A1A7YB09 A0A1U8DGC2 A0A151Z8P4 A0A3B5K2I8 A0A1U8DNQ3 A0A3B3B6H9 A0A3P8NDU6 A0A3Q3F6X1 A0A1S3F1D8 A0A3Q0SW69 A0A226P673 A0A384C0B7 G1L9G6 D2I143 H0ZIX2 A0A1A8KAV7 A0A3Q3SXK3 A0A091JKD9 A0A3Q1JFH6 A0A3Q1FCJ3
PDB
2HA8
E-value=1.67976e-47,
Score=474
Ontologies
GO
Topology
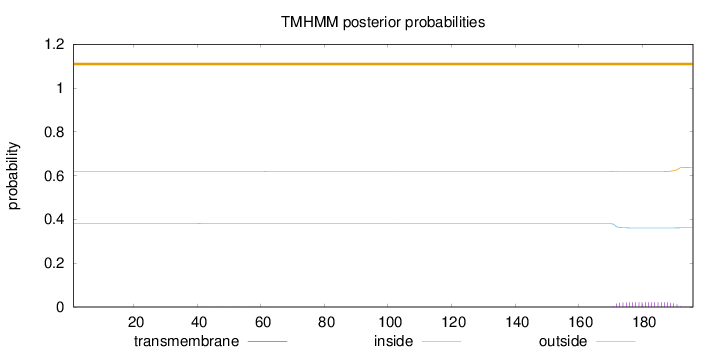
Length:
196
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.408
Exp number, first 60 AAs:
0.0043
Total prob of N-in:
0.38085
outside
1 - 196
Population Genetic Test Statistics
Pi
19.585466
Theta
21.930055
Tajima's D
-1.46214
CLR
0.538287
CSRT
0.0623468826558672
Interpretation
Uncertain
