Gene
KWMTBOMO07018
Pre Gene Modal
BGIBMGA010465
Annotation
PREDICTED:_neutral_ceramidase_[Amyelois_transitella]
Full name
Neutral ceramidase
Alternative Name
Neutral N-acylsphingosine amidohydrolase
Neutral acylsphingosine deacylase
Slug-a-bed protein
Acylsphingosine deacylase
N-acylsphingosine amidohydrolase
Neutral acylsphingosine deacylase
Slug-a-bed protein
Acylsphingosine deacylase
N-acylsphingosine amidohydrolase
Location in the cell
Lysosomal Reliability : 0.886
Sequence
CDS
ATGTTATTCGCGTGGTGCGTGCTTGCGTGCGTGACGGCGGCGGATGCGCTGCGTGTCGGAGCTGGCATAGCTGACGTCACTGGCCCACCGGCTGAAATTGCTTTTATGGGCTATGCTCAACTCGAGCAGATCGGGCATGGAATTCACCTGCGTCAGTTCTCAAGGGCCTTCGTGATAGAAGATAACAGTGGGGACACTGTTAAACGACTGGTCTTCGTGTCTGTTGATGCTGCGATGATGGGACATGGAGTTAGGAAAGAGGTAATAAGACGGCTGCAGAAGCGCTTTGGCGTAATCTACAACGAGGATAATGTGATCATCAGCGGCACCCACACTCACTCCACTCCTGGAGGATTTCTAATGGATTTCCTCTTCGATCTACCCATCCTTGGATTCGTGAAGGAGACGTACATAGCTTATGTCCTTGGAATTTATAAGAGCATAGTAATAGCTCACAGCAGGCTGACGTCGGCTCGGATAAAATACGGCGAGGCTGAACTTCTAGATGCGAATATCAACAGATCTCCGAAGTCATACTTGAACAATCCGGCAGAAGAAAGATCAAAATACAAATACGACGTGGATAAGACGTTGTCTCAAGTGAGATTCCTTGCAACGACGGGCGAGGTCATCGGCGTGATCAACTGGTTCGCCGTCCACCCGACCAGCATGAACAACACCAATAGGCTGGTGTCCTCGGACAACGTCGGATACGCTTCGCTACTCATGGAGAAGGCTCTGAATGGGAACAATACGTTACCGGGGAAAGGCGCAATAGTCTGCGCGTTCGGGTCAACGAACCTCGGCGACGTGTCCCCCAACACGCGCGGCCCGCGCTGCGAGCGGAGCGGCCGGCCGTGCGACCAGCAGGAGCAGCTCTGCGGGAAAAAGGAGCGCTGCTTCGCGTCCGGCCCGGGCAGGGACATGTTCGACAGCACTAGGATCATCGCTACCAAACTATTCGAAACTGCCATGAAAGTTCTCAGACAGCCCGGGGAAGATATAACTGGCACAGTGGCCGTGGCCCACCAATACGTCAAGATGCCAGATGAAGAAGTCCACCCATTCGATCCTGTGACAGAAACATTTAACATTAGTGTAAAAGTGAAGGGCTGTCTCCCGGCCATGGGGTATAGTTTCGCAGCAGGGACCACTGACGGGCCAGGGGCGTTTGACTTCAAACAGGGAACTAAAACATCTAACCCTCTCTGGAACGCCGTGAGGGACTTCATAGCCGAGCCGACGGAGGACGATGTACAGTGCCAAGCTCCAAAGCCCATCCTGCTGGCCACCGGAAGGGCTAAGTTCCCTTACGAGTGGCAGCCCCGGGTGGTGTCGTGCGCGGTCGCCAGGATCGGCTCGTTGTACTTGGCGGCCGTCCCCGGGGAGTTCACCACCATGTCTGGGAGGAGATTGCGGAACATCATCTCCAAGACCGCCCCACGACCCAACCGGGTCGTCATAGCCGGCCTCTCCAACATATACTCGGACTACGTGGCTACCCCTGAGGAGTACCAGGTGCAAAGGTACGAAGCAGCTTCAACCATCTACGGGCCCCACACATTGGACATCTACTTGAACAAATACCTTGAACTCACCGAGGCTCTACTCGAGAACAAAACCCCCGAGTCTGGTCCGGACCCTCCTGACTTGAGCGGCCAGCTGATCACTCTGGTGCCCCCCGTGCTGTGGGACTCCGCCCCCTGGAGGAAGGAGTTTGGAGACTGCGTGCAGCAACCGCAGTCCAGCTACAGCTACGGAGACGTCGTGGTAGCCGCTTTTGTGTCGGGACACCCACGGAACAGCGTCCGCCACGGCCGCTGGTACATGGCCGTAGAGAAACTCGAATCGGAGATCGATGACGCCTGGACTGTCATCGCGACTGATGCGGATTGGGAGACCAAATTCACCTGGCACCGTGACTCCAAAGTCTTGGGAACGAGTCACGTGGAACTCGAATGGGAGATACCCCCCGGGACTCCCCCCGGTACGTATCGGCTCCACCACTACGGGAACTACAAGTACGTTTTGGGTGGGATATACCCATACCACGGCTTCACTGATAGCTTCAAGGTCTCTTAA
Protein
MLFAWCVLACVTAADALRVGAGIADVTGPPAEIAFMGYAQLEQIGHGIHLRQFSRAFVIEDNSGDTVKRLVFVSVDAAMMGHGVRKEVIRRLQKRFGVIYNEDNVIISGTHTHSTPGGFLMDFLFDLPILGFVKETYIAYVLGIYKSIVIAHSRLTSARIKYGEAELLDANINRSPKSYLNNPAEERSKYKYDVDKTLSQVRFLATTGEVIGVINWFAVHPTSMNNTNRLVSSDNVGYASLLMEKALNGNNTLPGKGAIVCAFGSTNLGDVSPNTRGPRCERSGRPCDQQEQLCGKKERCFASGPGRDMFDSTRIIATKLFETAMKVLRQPGEDITGTVAVAHQYVKMPDEEVHPFDPVTETFNISVKVKGCLPAMGYSFAAGTTDGPGAFDFKQGTKTSNPLWNAVRDFIAEPTEDDVQCQAPKPILLATGRAKFPYEWQPRVVSCAVARIGSLYLAAVPGEFTTMSGRRLRNIISKTAPRPNRVVIAGLSNIYSDYVATPEEYQVQRYEAASTIYGPHTLDIYLNKYLELTEALLENKTPESGPDPPDLSGQLITLVPPVLWDSAPWRKEFGDCVQQPQSSYSYGDVVVAAFVSGHPRNSVRHGRWYMAVEKLESEIDDAWTVIATDADWETKFTWHRDSKVLGTSHVELEWEIPPGTPPGTYRLHHYGNYKYVLGGIYPYHGFTDSFKVS
Summary
Description
Hydrolyzes the sphingolipid ceramide into sphingosine and free fatty acid at an optimal pH of 6.5-7.5. Acts as a key regulator of sphingolipid signaling metabolites by generating sphingosine at the cell surface. Regulates synaptic vesicle exocytosis and trafficking by controlling presynaptic terminal sphingolipid composition.
Hydrolyzes the sphingolipid ceramide into sphingosine and free fatty acid at an optimal pH of 6.5-7.5. Acts as a key regulator of sphingolipid signaling metabolites by generating sphingosine at the cell surface (By similarity).
Hydrolyzes the sphingolipid ceramide into sphingosine and free fatty acid at an optimal pH of 6.5-7.5. Acts as a key regulator of sphingolipid signaling metabolites by generating sphingosine at the cell surface (By similarity).
Catalytic Activity
an N-acylsphing-4-enine + H2O = a fatty acid + sphing-4-enine
Miscellaneous
Overexpression rescues retinal degeneration in arrestin and phospholipase C mutants, probably by facilitating membrane turnover and endocytosis of rhodopsin in photoreceptors.
Similarity
Belongs to the neutral ceramidase family.
Keywords
Complete proteome
Glycoprotein
Hydrolase
Lipid metabolism
Reference proteome
Secreted
Signal
Sphingolipid metabolism
Feature
chain Neutral ceramidase
Uniprot
H9JLR3
A0A2H1VYE1
A0A2A4K9C0
A0A1E1WP24
A0A194PI22
A0A2W1B8Z5
+ More
A0A194QTJ8 A0A0L7LUS4 A0A212EIH2 A0A195DRJ6 A0A158NBE4 A0A1W4WKF1 A0A232F654 A0A195FCN6 K7ITX7 A0A026W3S6 A0A3L8DSF7 Q173S2 A0A1S4FFJ2 A0A154P9R8 A0A2K8JRS6 A0A084VN16 A0A023F618 A0A224XFS4 B0WGE6 A0A0P4VKF0 A0A182U547 A0A0C9RVC1 A0A182G996 A0A1I8PNB8 A0A1Q3FNI5 A0A1Q3FNN9 A0A182Y541 A0A1Q3FNZ9 A0A182QIH4 A0A182RL44 A0A1Q3FNM7 D6X3Y4 A0A1L8DPV5 A0A1L8EGU5 A0A1B0CQR1 A0A1L8EGN3 T1PE63 Q7QI06 A0A182PPX0 E9IBK4 A0A182W445 A0A0L0BLG6 A0A1A9ZZK7 I2CMG2 A0A067RJZ6 A0A182FIG3 W5JJX8 A0A1L8DQ15 V5GV97 A0A2M4ATB9 A0A1B0FRH6 A0A182JG20 R4N4U2 A0A034V4F0 A0A0K8USE8 A0A1A9WN33 A0A182MZX1 A0A2M4BFF7 A0A146ME47 A0A0A9W5T8 B4K8R6 A0A0M4EGS5 A0A1A9XTC7 B4LYM3 A0A182US50 A0A0A1WH00 D6X3Y5 B4JVA2 A0A0A1WKA2 B4R1M6 B4HZR3 B3P584 A4V3N7 B4PLW9 Q9VA70 A0A0R3NKD0 Q29C43 B4NGW1 W8BUB2 A0A3B0K2B9 A0A3B0JCJ4 A0A3B0K988 A0A1W4VRB3 A0A336ML87 A0A182IDP2 A0A182XNH7 A0A0A9WE73 A0A1J1J8W7 A0A1Q3FFP1 A0A1Q3FFN4 A0A1B0BUR0 B3MTB9 A0A182U2Y6 A0A182UYT8
A0A194QTJ8 A0A0L7LUS4 A0A212EIH2 A0A195DRJ6 A0A158NBE4 A0A1W4WKF1 A0A232F654 A0A195FCN6 K7ITX7 A0A026W3S6 A0A3L8DSF7 Q173S2 A0A1S4FFJ2 A0A154P9R8 A0A2K8JRS6 A0A084VN16 A0A023F618 A0A224XFS4 B0WGE6 A0A0P4VKF0 A0A182U547 A0A0C9RVC1 A0A182G996 A0A1I8PNB8 A0A1Q3FNI5 A0A1Q3FNN9 A0A182Y541 A0A1Q3FNZ9 A0A182QIH4 A0A182RL44 A0A1Q3FNM7 D6X3Y4 A0A1L8DPV5 A0A1L8EGU5 A0A1B0CQR1 A0A1L8EGN3 T1PE63 Q7QI06 A0A182PPX0 E9IBK4 A0A182W445 A0A0L0BLG6 A0A1A9ZZK7 I2CMG2 A0A067RJZ6 A0A182FIG3 W5JJX8 A0A1L8DQ15 V5GV97 A0A2M4ATB9 A0A1B0FRH6 A0A182JG20 R4N4U2 A0A034V4F0 A0A0K8USE8 A0A1A9WN33 A0A182MZX1 A0A2M4BFF7 A0A146ME47 A0A0A9W5T8 B4K8R6 A0A0M4EGS5 A0A1A9XTC7 B4LYM3 A0A182US50 A0A0A1WH00 D6X3Y5 B4JVA2 A0A0A1WKA2 B4R1M6 B4HZR3 B3P584 A4V3N7 B4PLW9 Q9VA70 A0A0R3NKD0 Q29C43 B4NGW1 W8BUB2 A0A3B0K2B9 A0A3B0JCJ4 A0A3B0K988 A0A1W4VRB3 A0A336ML87 A0A182IDP2 A0A182XNH7 A0A0A9WE73 A0A1J1J8W7 A0A1Q3FFP1 A0A1Q3FFN4 A0A1B0BUR0 B3MTB9 A0A182U2Y6 A0A182UYT8
EC Number
3.5.1.23
Pubmed
19121390
26354079
28756777
26227816
22118469
21347285
+ More
28648823 20075255 24508170 30249741 17510324 24438588 25474469 27129103 26483478 25244985 18362917 19820115 25315136 12364791 14747013 17210077 21282665 26108605 22517621 24845553 20920257 23761445 23601004 25348373 26823975 25401762 17994087 18057021 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12153720 12637747 15356190 14769922 15632085 24495485
28648823 20075255 24508170 30249741 17510324 24438588 25474469 27129103 26483478 25244985 18362917 19820115 25315136 12364791 14747013 17210077 21282665 26108605 22517621 24845553 20920257 23761445 23601004 25348373 26823975 25401762 17994087 18057021 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12153720 12637747 15356190 14769922 15632085 24495485
EMBL
BABH01005208
BABH01005209
BABH01005210
ODYU01005218
SOQ45860.1
NWSH01000039
+ More
PCG80383.1 GDQN01002261 JAT88793.1 KQ459603 KPI92967.1 KZ150341 PZC71311.1 KQ461194 KPJ06871.1 JTDY01000041 KOB79228.1 AGBW02014643 OWR41277.1 KQ980581 KYN15461.1 ADTU01011000 ADTU01011001 ADTU01011002 ADTU01011003 ADTU01011004 ADTU01011005 ADTU01011006 ADTU01011007 ADTU01011008 ADTU01011009 NNAY01000885 OXU26062.1 KQ981673 KYN38173.1 KK107453 EZA50648.1 QOIP01000004 RLU23242.1 CH477417 EAT41313.1 KQ434839 KZC07968.1 KY030989 ATU82740.1 ATLV01014673 KE524978 KFB39360.1 GBBI01002065 JAC16647.1 GFTR01007768 JAW08658.1 DS231926 EDS26943.1 GDKW01001794 JAI54801.1 GBYB01011646 JAG81413.1 JXUM01049339 KQ561600 KXJ78038.1 GFDL01005885 JAV29160.1 GFDL01005826 JAV29219.1 GFDL01005839 JAV29206.1 AXCN02002023 GFDL01005878 JAV29167.1 KQ971379 EEZ97353.1 GFDF01005592 JAV08492.1 GFDG01000986 JAV17813.1 AJWK01023885 AJWK01023886 AJWK01023887 GFDG01000985 JAV17814.1 KA646243 AFP60872.1 AAAB01008811 EAA04917.3 GL762111 EFZ22084.1 JRES01001702 KNC20803.1 JQ308540 AFJ68095.1 KK852425 KDR24151.1 ADMH02000912 ETN64687.1 GFDF01005593 JAV08491.1 GALX01004308 JAB64158.1 GGFK01010693 MBW44014.1 CCAG010022123 JX569799 AGL39308.1 GAKP01021563 JAC37389.1 GDHF01022808 GDHF01014766 GDHF01001327 JAI29506.1 JAI37548.1 JAI50987.1 GGFJ01002644 MBW51785.1 GDHC01001347 JAQ17282.1 GBHO01040465 GBRD01013004 GBRD01013003 JAG03139.1 JAG52822.1 CH933806 EDW15485.1 KRG01581.1 CP012526 ALC47628.1 CH940650 EDW68043.1 KRF83622.1 GBXI01016527 JAC97764.1 EEZ97352.2 CH916374 EDV91422.1 GBXI01015337 JAC98954.1 CM000364 EDX15014.1 CH480819 EDW53520.1 CH954182 EDV53067.1 AE014297 BT150409 AAF57052.1 AAN14232.1 AAN14233.1 AAN14234.1 AGB96488.1 AHA95713.1 CM000160 EDW99106.1 KRK04661.1 KRK04662.1 AB112076 BT004471 CM000070 KRS99567.1 CH964272 EDW84458.1 GAMC01013846 GAMC01013845 JAB92709.1 OUUW01000005 SPP80099.1 SPP80097.1 SPP80098.1 UFQT01001467 SSX30700.1 APCN01004753 GBHO01040464 GBRD01013007 GBRD01013005 JAG03140.1 JAG52819.1 CVRI01000074 CRL07958.1 GFDL01008667 JAV26378.1 GFDL01008699 JAV26346.1 JXJN01020882 JXJN01020883 CH902623 EDV30509.1 KPU72902.1
PCG80383.1 GDQN01002261 JAT88793.1 KQ459603 KPI92967.1 KZ150341 PZC71311.1 KQ461194 KPJ06871.1 JTDY01000041 KOB79228.1 AGBW02014643 OWR41277.1 KQ980581 KYN15461.1 ADTU01011000 ADTU01011001 ADTU01011002 ADTU01011003 ADTU01011004 ADTU01011005 ADTU01011006 ADTU01011007 ADTU01011008 ADTU01011009 NNAY01000885 OXU26062.1 KQ981673 KYN38173.1 KK107453 EZA50648.1 QOIP01000004 RLU23242.1 CH477417 EAT41313.1 KQ434839 KZC07968.1 KY030989 ATU82740.1 ATLV01014673 KE524978 KFB39360.1 GBBI01002065 JAC16647.1 GFTR01007768 JAW08658.1 DS231926 EDS26943.1 GDKW01001794 JAI54801.1 GBYB01011646 JAG81413.1 JXUM01049339 KQ561600 KXJ78038.1 GFDL01005885 JAV29160.1 GFDL01005826 JAV29219.1 GFDL01005839 JAV29206.1 AXCN02002023 GFDL01005878 JAV29167.1 KQ971379 EEZ97353.1 GFDF01005592 JAV08492.1 GFDG01000986 JAV17813.1 AJWK01023885 AJWK01023886 AJWK01023887 GFDG01000985 JAV17814.1 KA646243 AFP60872.1 AAAB01008811 EAA04917.3 GL762111 EFZ22084.1 JRES01001702 KNC20803.1 JQ308540 AFJ68095.1 KK852425 KDR24151.1 ADMH02000912 ETN64687.1 GFDF01005593 JAV08491.1 GALX01004308 JAB64158.1 GGFK01010693 MBW44014.1 CCAG010022123 JX569799 AGL39308.1 GAKP01021563 JAC37389.1 GDHF01022808 GDHF01014766 GDHF01001327 JAI29506.1 JAI37548.1 JAI50987.1 GGFJ01002644 MBW51785.1 GDHC01001347 JAQ17282.1 GBHO01040465 GBRD01013004 GBRD01013003 JAG03139.1 JAG52822.1 CH933806 EDW15485.1 KRG01581.1 CP012526 ALC47628.1 CH940650 EDW68043.1 KRF83622.1 GBXI01016527 JAC97764.1 EEZ97352.2 CH916374 EDV91422.1 GBXI01015337 JAC98954.1 CM000364 EDX15014.1 CH480819 EDW53520.1 CH954182 EDV53067.1 AE014297 BT150409 AAF57052.1 AAN14232.1 AAN14233.1 AAN14234.1 AGB96488.1 AHA95713.1 CM000160 EDW99106.1 KRK04661.1 KRK04662.1 AB112076 BT004471 CM000070 KRS99567.1 CH964272 EDW84458.1 GAMC01013846 GAMC01013845 JAB92709.1 OUUW01000005 SPP80099.1 SPP80097.1 SPP80098.1 UFQT01001467 SSX30700.1 APCN01004753 GBHO01040464 GBRD01013007 GBRD01013005 JAG03140.1 JAG52819.1 CVRI01000074 CRL07958.1 GFDL01008667 JAV26378.1 GFDL01008699 JAV26346.1 JXJN01020882 JXJN01020883 CH902623 EDV30509.1 KPU72902.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000078492 UP000005205 UP000192223 UP000215335 UP000078541 UP000002358 UP000053097 UP000279307 UP000008820 UP000076502 UP000030765 UP000002320 UP000075902 UP000069940 UP000249989 UP000095300 UP000076408 UP000075886 UP000075900 UP000007266 UP000092461 UP000095301 UP000007062 UP000075885 UP000075920 UP000037069 UP000092445 UP000027135 UP000069272 UP000000673 UP000092444 UP000075880 UP000091820 UP000075884 UP000009192 UP000092553 UP000092443 UP000008792 UP000075903 UP000001070 UP000000304 UP000001292 UP000008711 UP000000803 UP000002282 UP000001819 UP000007798 UP000268350 UP000192221 UP000075840 UP000076407 UP000183832 UP000092460 UP000007801
UP000078492 UP000005205 UP000192223 UP000215335 UP000078541 UP000002358 UP000053097 UP000279307 UP000008820 UP000076502 UP000030765 UP000002320 UP000075902 UP000069940 UP000249989 UP000095300 UP000076408 UP000075886 UP000075900 UP000007266 UP000092461 UP000095301 UP000007062 UP000075885 UP000075920 UP000037069 UP000092445 UP000027135 UP000069272 UP000000673 UP000092444 UP000075880 UP000091820 UP000075884 UP000009192 UP000092553 UP000092443 UP000008792 UP000075903 UP000001070 UP000000304 UP000001292 UP000008711 UP000000803 UP000002282 UP000001819 UP000007798 UP000268350 UP000192221 UP000075840 UP000076407 UP000183832 UP000092460 UP000007801
Interpro
SUPFAM
SSF54160
SSF54160
Gene 3D
CDD
ProteinModelPortal
H9JLR3
A0A2H1VYE1
A0A2A4K9C0
A0A1E1WP24
A0A194PI22
A0A2W1B8Z5
+ More
A0A194QTJ8 A0A0L7LUS4 A0A212EIH2 A0A195DRJ6 A0A158NBE4 A0A1W4WKF1 A0A232F654 A0A195FCN6 K7ITX7 A0A026W3S6 A0A3L8DSF7 Q173S2 A0A1S4FFJ2 A0A154P9R8 A0A2K8JRS6 A0A084VN16 A0A023F618 A0A224XFS4 B0WGE6 A0A0P4VKF0 A0A182U547 A0A0C9RVC1 A0A182G996 A0A1I8PNB8 A0A1Q3FNI5 A0A1Q3FNN9 A0A182Y541 A0A1Q3FNZ9 A0A182QIH4 A0A182RL44 A0A1Q3FNM7 D6X3Y4 A0A1L8DPV5 A0A1L8EGU5 A0A1B0CQR1 A0A1L8EGN3 T1PE63 Q7QI06 A0A182PPX0 E9IBK4 A0A182W445 A0A0L0BLG6 A0A1A9ZZK7 I2CMG2 A0A067RJZ6 A0A182FIG3 W5JJX8 A0A1L8DQ15 V5GV97 A0A2M4ATB9 A0A1B0FRH6 A0A182JG20 R4N4U2 A0A034V4F0 A0A0K8USE8 A0A1A9WN33 A0A182MZX1 A0A2M4BFF7 A0A146ME47 A0A0A9W5T8 B4K8R6 A0A0M4EGS5 A0A1A9XTC7 B4LYM3 A0A182US50 A0A0A1WH00 D6X3Y5 B4JVA2 A0A0A1WKA2 B4R1M6 B4HZR3 B3P584 A4V3N7 B4PLW9 Q9VA70 A0A0R3NKD0 Q29C43 B4NGW1 W8BUB2 A0A3B0K2B9 A0A3B0JCJ4 A0A3B0K988 A0A1W4VRB3 A0A336ML87 A0A182IDP2 A0A182XNH7 A0A0A9WE73 A0A1J1J8W7 A0A1Q3FFP1 A0A1Q3FFN4 A0A1B0BUR0 B3MTB9 A0A182U2Y6 A0A182UYT8
A0A194QTJ8 A0A0L7LUS4 A0A212EIH2 A0A195DRJ6 A0A158NBE4 A0A1W4WKF1 A0A232F654 A0A195FCN6 K7ITX7 A0A026W3S6 A0A3L8DSF7 Q173S2 A0A1S4FFJ2 A0A154P9R8 A0A2K8JRS6 A0A084VN16 A0A023F618 A0A224XFS4 B0WGE6 A0A0P4VKF0 A0A182U547 A0A0C9RVC1 A0A182G996 A0A1I8PNB8 A0A1Q3FNI5 A0A1Q3FNN9 A0A182Y541 A0A1Q3FNZ9 A0A182QIH4 A0A182RL44 A0A1Q3FNM7 D6X3Y4 A0A1L8DPV5 A0A1L8EGU5 A0A1B0CQR1 A0A1L8EGN3 T1PE63 Q7QI06 A0A182PPX0 E9IBK4 A0A182W445 A0A0L0BLG6 A0A1A9ZZK7 I2CMG2 A0A067RJZ6 A0A182FIG3 W5JJX8 A0A1L8DQ15 V5GV97 A0A2M4ATB9 A0A1B0FRH6 A0A182JG20 R4N4U2 A0A034V4F0 A0A0K8USE8 A0A1A9WN33 A0A182MZX1 A0A2M4BFF7 A0A146ME47 A0A0A9W5T8 B4K8R6 A0A0M4EGS5 A0A1A9XTC7 B4LYM3 A0A182US50 A0A0A1WH00 D6X3Y5 B4JVA2 A0A0A1WKA2 B4R1M6 B4HZR3 B3P584 A4V3N7 B4PLW9 Q9VA70 A0A0R3NKD0 Q29C43 B4NGW1 W8BUB2 A0A3B0K2B9 A0A3B0JCJ4 A0A3B0K988 A0A1W4VRB3 A0A336ML87 A0A182IDP2 A0A182XNH7 A0A0A9WE73 A0A1J1J8W7 A0A1Q3FFP1 A0A1Q3FFN4 A0A1B0BUR0 B3MTB9 A0A182U2Y6 A0A182UYT8
PDB
4WGK
E-value=7.99273e-159,
Score=1440
Ontologies
PATHWAY
GO
PANTHER
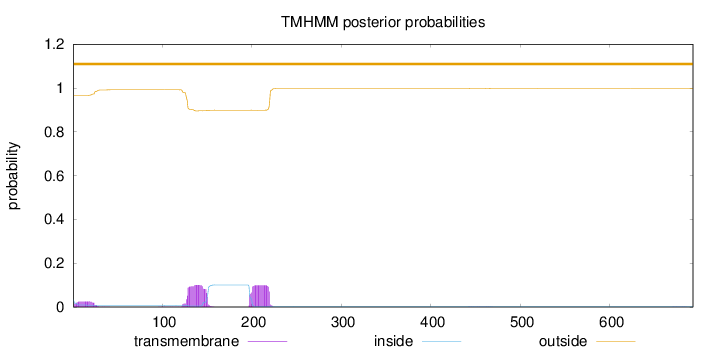
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.966831

Length:
693
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.98984999999999
Exp number, first 60 AAs:
0.5606
Total prob of N-in:
0.03359
outside
1 - 693
Population Genetic Test Statistics
Pi
208.130216
Theta
16.09512
Tajima's D
0.966313
CLR
1.035918
CSRT
0.658317084145793
Interpretation
Uncertain
