Gene
KWMTBOMO07017
Pre Gene Modal
BGIBMGA010466
Annotation
PREDICTED:_neutral_ceramidase-like_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 1.326
Sequence
CDS
ATGGCATTATCTTTTGTGGGAAATGTCGTGATTACCGTGATCGTATTGGGCGTAATCAGTGGCACGAGTGCTACTTTCACTTTTAAAACCAAGGACAATATTGATAATCATGAAGATGATCAGTCTAAGCAGTTTCCGAGGCCAGAGCCAGAGGCGGCTTCGTCTTACGAAGTCGGCGTTGGTATTGCAGATATGACCGGACCCTGTGTTGAAATAACATTTATGGGTTACGCTGAGCTCGGACAGTCTGGCGAAGGTATACATTTGAGGCAGTTTTCCAGAGCCTTCATATTTTCGAAGGGGAATACCAGAGTGGTACTAGTCACAGCTGAAGTGCAGGCAGTAGGCATTGCCGTCAGGAGACAGGTGGTGAAGAATTTGCAAGAGCGTTACGGTGACATGTACAGCCTCCGCAACGTGATAATCACTGGAACGCACACGCACAGCACCCCTGGTGGATATCTGGTGGATTTCATTCTGGACGTTAGCATATTAGGATTCTCTAAGGAGACTTTCAATGCCTACGTTGACGGAATTACTAGGAGCATAGTCCGCGCCCATGAAAACATGGTACCAGCGCGTTTGGTGTTCGCTCAGACGAAAGTAAAGAACGCCCACAAGAACCGATCTCCCTACTCCTATAATAACAATCCTGCTGAGGAACGAGCTAGCCCACCTGGTGTTAAGAGGTAA
Protein
MALSFVGNVVITVIVLGVISGTSATFTFKTKDNIDNHEDDQSKQFPRPEPEAASSYEVGVGIADMTGPCVEITFMGYAELGQSGEGIHLRQFSRAFIFSKGNTRVVLVTAEVQAVGIAVRRQVVKNLQERYGDMYSLRNVIITGTHTHSTPGGYLVDFILDVSILGFSKETFNAYVDGITRSIVRAHENMVPARLVFAQTKVKNAHKNRSPYSYNNNPAEERASPPGVKR
Summary
Uniprot
H9JLR4
A0A2A4K9P2
A0A1E1VZH0
A0A2W1BEQ2
A0A2H1VBN7
A0A3S2LIR5
+ More
A0A194QND7 A0A194PJR3 A0A0L7LV17 A0A212EIG5 V5GRC4 A0A194QTJ8 A0A1E1WP24 A0A194PI22 A0A2H1VZ93 A0A182G996 A0A2W1B8Z5 A0A2H1VYE1 V5GV97 A0A2A4K9C0 A0A087ZNN1 Q173S2 A0A1S4FFJ2 Q173S3 A0A0L7RCF7 A0A084VN16 B0XLL1 A0A182QIH4 A0A1Q3FFP1 W5JJX8 A0A182RL44 A0A0J7KWV6 A0A182Y541 A0A182LIT5 A0A1Q3FFN4 A0A182MJ71 A0A182W445 A0A182PPX0 Q7QI06 A0A182JG20 A0A2P8Y9J9 A0A1B0CQR1 A0A182IDP2 A0A182US50 D6X3Y5 A0A182XNH7 E2ADY5 A0A182U547 A0A2K8JRS6 A0A2M4ATB9 A0A1L8DPV5 B0WGE6 A0A023F618 A0A224XFS4 A0A146ME47 A0A0A9W5T8 A0A182MZX1 A0A1Q3FNN9 A0A1L8DQ15 A0A1Q3FNI5 A0A1Q3FNM7 A0A1Q3FNZ9 A0A182FIG3 A0A336ML87 R4N4U2 N6TBZ0 D6X3Y4 N6U9H2 U4TTS5 A0A232F654 A0A0M8ZWP9 A0A195BAE0 A0A182PKG1 T1HX07 A0A154P9R8 K7ITX7 A0A2M4BFF7 F4X8U0 A0A034V4F0 A0A2M4ACS0 A0A2M4ACS3 A0A0P4VKF0 T1PE63 A0A1I8PNB8 A0A2M4BF47 A0A195DRJ6 E9IRA3 A0A182I907 A0A2M4ADN9 A0A151I6X0 A0A182UYT8 A0A182LAZ3 A0A182U2Y6 A0A182WVC6 A0A2M4BF41 A0A0K8USE8 A0A195FCN6 A0A0L0BLG6 Q7QG18
A0A194QND7 A0A194PJR3 A0A0L7LV17 A0A212EIG5 V5GRC4 A0A194QTJ8 A0A1E1WP24 A0A194PI22 A0A2H1VZ93 A0A182G996 A0A2W1B8Z5 A0A2H1VYE1 V5GV97 A0A2A4K9C0 A0A087ZNN1 Q173S2 A0A1S4FFJ2 Q173S3 A0A0L7RCF7 A0A084VN16 B0XLL1 A0A182QIH4 A0A1Q3FFP1 W5JJX8 A0A182RL44 A0A0J7KWV6 A0A182Y541 A0A182LIT5 A0A1Q3FFN4 A0A182MJ71 A0A182W445 A0A182PPX0 Q7QI06 A0A182JG20 A0A2P8Y9J9 A0A1B0CQR1 A0A182IDP2 A0A182US50 D6X3Y5 A0A182XNH7 E2ADY5 A0A182U547 A0A2K8JRS6 A0A2M4ATB9 A0A1L8DPV5 B0WGE6 A0A023F618 A0A224XFS4 A0A146ME47 A0A0A9W5T8 A0A182MZX1 A0A1Q3FNN9 A0A1L8DQ15 A0A1Q3FNI5 A0A1Q3FNM7 A0A1Q3FNZ9 A0A182FIG3 A0A336ML87 R4N4U2 N6TBZ0 D6X3Y4 N6U9H2 U4TTS5 A0A232F654 A0A0M8ZWP9 A0A195BAE0 A0A182PKG1 T1HX07 A0A154P9R8 K7ITX7 A0A2M4BFF7 F4X8U0 A0A034V4F0 A0A2M4ACS0 A0A2M4ACS3 A0A0P4VKF0 T1PE63 A0A1I8PNB8 A0A2M4BF47 A0A195DRJ6 E9IRA3 A0A182I907 A0A2M4ADN9 A0A151I6X0 A0A182UYT8 A0A182LAZ3 A0A182U2Y6 A0A182WVC6 A0A2M4BF41 A0A0K8USE8 A0A195FCN6 A0A0L0BLG6 Q7QG18
Pubmed
EMBL
BABH01005206
BABH01005207
BABH01005208
NWSH01000039
PCG80382.1
GDQN01010918
+ More
JAT80136.1 KZ150341 PZC71310.1 ODYU01001690 SOQ38260.1 RSAL01000003 RVE54728.1 KQ461194 KPJ06869.1 KQ459603 KPI92969.1 JTDY01000041 KOB79229.1 AGBW02014643 OWR41276.1 GALX01004309 JAB64157.1 KPJ06871.1 GDQN01002261 JAT88793.1 KPI92967.1 ODYU01005219 SOQ45862.1 JXUM01049339 KQ561600 KXJ78038.1 PZC71311.1 ODYU01005218 SOQ45860.1 GALX01004308 JAB64158.1 PCG80383.1 CH477417 EAT41313.1 EAT41312.1 KQ414616 KOC68485.1 ATLV01014673 KE524978 KFB39360.1 DS234534 EDS34360.1 AXCN02002023 GFDL01008667 JAV26378.1 ADMH02000912 ETN64687.1 LBMM01002475 KMQ94783.1 GFDL01008699 JAV26346.1 AXCM01006278 AAAB01008811 EAA04917.3 PYGN01000778 PSN40922.1 AJWK01023885 AJWK01023886 AJWK01023887 APCN01004753 KQ971379 EEZ97352.2 GL438827 EFN68218.1 KY030989 ATU82740.1 GGFK01010693 MBW44014.1 GFDF01005592 JAV08492.1 DS231926 EDS26943.1 GBBI01002065 JAC16647.1 GFTR01007768 JAW08658.1 GDHC01001347 JAQ17282.1 GBHO01040465 GBRD01013004 GBRD01013003 JAG03139.1 JAG52822.1 GFDL01005826 JAV29219.1 GFDF01005593 JAV08491.1 GFDL01005885 JAV29160.1 GFDL01005878 JAV29167.1 GFDL01005839 JAV29206.1 UFQT01001467 SSX30700.1 JX569799 AGL39308.1 APGK01043745 APGK01043746 APGK01043747 APGK01043748 APGK01043749 APGK01043750 APGK01043751 APGK01043752 APGK01043753 KB741017 ENN75248.1 EEZ97353.1 ENN75247.1 KB631644 ERL84934.1 NNAY01000885 OXU26062.1 KQ435847 KOX71109.1 KQ976540 KYM81192.1 ACPB03025916 KQ434839 KZC07968.1 GGFJ01002644 MBW51785.1 GL888932 EGI57359.1 GAKP01021563 JAC37389.1 GGFK01005263 MBW38584.1 GGFK01005262 MBW38583.1 GDKW01001794 JAI54801.1 KA646243 AFP60872.1 GGFJ01002534 MBW51675.1 KQ980581 KYN15461.1 GL765107 EFZ16859.1 APCN01003147 GGFK01005584 MBW38905.1 KQ978455 KYM93916.1 GGFJ01002535 MBW51676.1 GDHF01022808 GDHF01014766 GDHF01001327 JAI29506.1 JAI37548.1 JAI50987.1 KQ981673 KYN38173.1 JRES01001702 KNC20803.1 AAAB01008844 EAA05985.5
JAT80136.1 KZ150341 PZC71310.1 ODYU01001690 SOQ38260.1 RSAL01000003 RVE54728.1 KQ461194 KPJ06869.1 KQ459603 KPI92969.1 JTDY01000041 KOB79229.1 AGBW02014643 OWR41276.1 GALX01004309 JAB64157.1 KPJ06871.1 GDQN01002261 JAT88793.1 KPI92967.1 ODYU01005219 SOQ45862.1 JXUM01049339 KQ561600 KXJ78038.1 PZC71311.1 ODYU01005218 SOQ45860.1 GALX01004308 JAB64158.1 PCG80383.1 CH477417 EAT41313.1 EAT41312.1 KQ414616 KOC68485.1 ATLV01014673 KE524978 KFB39360.1 DS234534 EDS34360.1 AXCN02002023 GFDL01008667 JAV26378.1 ADMH02000912 ETN64687.1 LBMM01002475 KMQ94783.1 GFDL01008699 JAV26346.1 AXCM01006278 AAAB01008811 EAA04917.3 PYGN01000778 PSN40922.1 AJWK01023885 AJWK01023886 AJWK01023887 APCN01004753 KQ971379 EEZ97352.2 GL438827 EFN68218.1 KY030989 ATU82740.1 GGFK01010693 MBW44014.1 GFDF01005592 JAV08492.1 DS231926 EDS26943.1 GBBI01002065 JAC16647.1 GFTR01007768 JAW08658.1 GDHC01001347 JAQ17282.1 GBHO01040465 GBRD01013004 GBRD01013003 JAG03139.1 JAG52822.1 GFDL01005826 JAV29219.1 GFDF01005593 JAV08491.1 GFDL01005885 JAV29160.1 GFDL01005878 JAV29167.1 GFDL01005839 JAV29206.1 UFQT01001467 SSX30700.1 JX569799 AGL39308.1 APGK01043745 APGK01043746 APGK01043747 APGK01043748 APGK01043749 APGK01043750 APGK01043751 APGK01043752 APGK01043753 KB741017 ENN75248.1 EEZ97353.1 ENN75247.1 KB631644 ERL84934.1 NNAY01000885 OXU26062.1 KQ435847 KOX71109.1 KQ976540 KYM81192.1 ACPB03025916 KQ434839 KZC07968.1 GGFJ01002644 MBW51785.1 GL888932 EGI57359.1 GAKP01021563 JAC37389.1 GGFK01005263 MBW38584.1 GGFK01005262 MBW38583.1 GDKW01001794 JAI54801.1 KA646243 AFP60872.1 GGFJ01002534 MBW51675.1 KQ980581 KYN15461.1 GL765107 EFZ16859.1 APCN01003147 GGFK01005584 MBW38905.1 KQ978455 KYM93916.1 GGFJ01002535 MBW51676.1 GDHF01022808 GDHF01014766 GDHF01001327 JAI29506.1 JAI37548.1 JAI50987.1 KQ981673 KYN38173.1 JRES01001702 KNC20803.1 AAAB01008844 EAA05985.5
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000007151 UP000069940 UP000249989 UP000005203 UP000008820 UP000053825 UP000030765 UP000002320 UP000075886 UP000000673 UP000075900 UP000036403 UP000076408 UP000075882 UP000075883 UP000075920 UP000075885 UP000007062 UP000075880 UP000245037 UP000092461 UP000075840 UP000075903 UP000007266 UP000076407 UP000000311 UP000075902 UP000075884 UP000069272 UP000019118 UP000030742 UP000215335 UP000053105 UP000078540 UP000015103 UP000076502 UP000002358 UP000007755 UP000095301 UP000095300 UP000078492 UP000078542 UP000078541 UP000037069
UP000007151 UP000069940 UP000249989 UP000005203 UP000008820 UP000053825 UP000030765 UP000002320 UP000075886 UP000000673 UP000075900 UP000036403 UP000076408 UP000075882 UP000075883 UP000075920 UP000075885 UP000007062 UP000075880 UP000245037 UP000092461 UP000075840 UP000075903 UP000007266 UP000076407 UP000000311 UP000075902 UP000075884 UP000069272 UP000019118 UP000030742 UP000215335 UP000053105 UP000078540 UP000015103 UP000076502 UP000002358 UP000007755 UP000095301 UP000095300 UP000078492 UP000078542 UP000078541 UP000037069
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JLR4
A0A2A4K9P2
A0A1E1VZH0
A0A2W1BEQ2
A0A2H1VBN7
A0A3S2LIR5
+ More
A0A194QND7 A0A194PJR3 A0A0L7LV17 A0A212EIG5 V5GRC4 A0A194QTJ8 A0A1E1WP24 A0A194PI22 A0A2H1VZ93 A0A182G996 A0A2W1B8Z5 A0A2H1VYE1 V5GV97 A0A2A4K9C0 A0A087ZNN1 Q173S2 A0A1S4FFJ2 Q173S3 A0A0L7RCF7 A0A084VN16 B0XLL1 A0A182QIH4 A0A1Q3FFP1 W5JJX8 A0A182RL44 A0A0J7KWV6 A0A182Y541 A0A182LIT5 A0A1Q3FFN4 A0A182MJ71 A0A182W445 A0A182PPX0 Q7QI06 A0A182JG20 A0A2P8Y9J9 A0A1B0CQR1 A0A182IDP2 A0A182US50 D6X3Y5 A0A182XNH7 E2ADY5 A0A182U547 A0A2K8JRS6 A0A2M4ATB9 A0A1L8DPV5 B0WGE6 A0A023F618 A0A224XFS4 A0A146ME47 A0A0A9W5T8 A0A182MZX1 A0A1Q3FNN9 A0A1L8DQ15 A0A1Q3FNI5 A0A1Q3FNM7 A0A1Q3FNZ9 A0A182FIG3 A0A336ML87 R4N4U2 N6TBZ0 D6X3Y4 N6U9H2 U4TTS5 A0A232F654 A0A0M8ZWP9 A0A195BAE0 A0A182PKG1 T1HX07 A0A154P9R8 K7ITX7 A0A2M4BFF7 F4X8U0 A0A034V4F0 A0A2M4ACS0 A0A2M4ACS3 A0A0P4VKF0 T1PE63 A0A1I8PNB8 A0A2M4BF47 A0A195DRJ6 E9IRA3 A0A182I907 A0A2M4ADN9 A0A151I6X0 A0A182UYT8 A0A182LAZ3 A0A182U2Y6 A0A182WVC6 A0A2M4BF41 A0A0K8USE8 A0A195FCN6 A0A0L0BLG6 Q7QG18
A0A194QND7 A0A194PJR3 A0A0L7LV17 A0A212EIG5 V5GRC4 A0A194QTJ8 A0A1E1WP24 A0A194PI22 A0A2H1VZ93 A0A182G996 A0A2W1B8Z5 A0A2H1VYE1 V5GV97 A0A2A4K9C0 A0A087ZNN1 Q173S2 A0A1S4FFJ2 Q173S3 A0A0L7RCF7 A0A084VN16 B0XLL1 A0A182QIH4 A0A1Q3FFP1 W5JJX8 A0A182RL44 A0A0J7KWV6 A0A182Y541 A0A182LIT5 A0A1Q3FFN4 A0A182MJ71 A0A182W445 A0A182PPX0 Q7QI06 A0A182JG20 A0A2P8Y9J9 A0A1B0CQR1 A0A182IDP2 A0A182US50 D6X3Y5 A0A182XNH7 E2ADY5 A0A182U547 A0A2K8JRS6 A0A2M4ATB9 A0A1L8DPV5 B0WGE6 A0A023F618 A0A224XFS4 A0A146ME47 A0A0A9W5T8 A0A182MZX1 A0A1Q3FNN9 A0A1L8DQ15 A0A1Q3FNI5 A0A1Q3FNM7 A0A1Q3FNZ9 A0A182FIG3 A0A336ML87 R4N4U2 N6TBZ0 D6X3Y4 N6U9H2 U4TTS5 A0A232F654 A0A0M8ZWP9 A0A195BAE0 A0A182PKG1 T1HX07 A0A154P9R8 K7ITX7 A0A2M4BFF7 F4X8U0 A0A034V4F0 A0A2M4ACS0 A0A2M4ACS3 A0A0P4VKF0 T1PE63 A0A1I8PNB8 A0A2M4BF47 A0A195DRJ6 E9IRA3 A0A182I907 A0A2M4ADN9 A0A151I6X0 A0A182UYT8 A0A182LAZ3 A0A182U2Y6 A0A182WVC6 A0A2M4BF41 A0A0K8USE8 A0A195FCN6 A0A0L0BLG6 Q7QG18
PDB
4WGK
E-value=5.14812e-35,
Score=367
Ontologies
PATHWAY
PANTHER
Topology
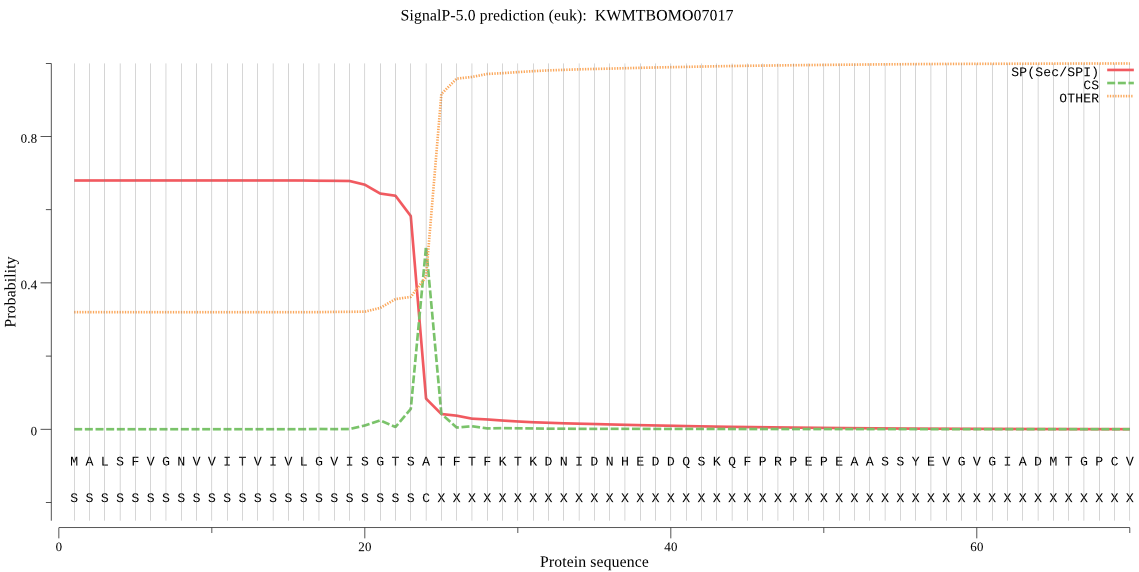
SignalP
Position: 1 - 24,
Likelihood: 0.680466
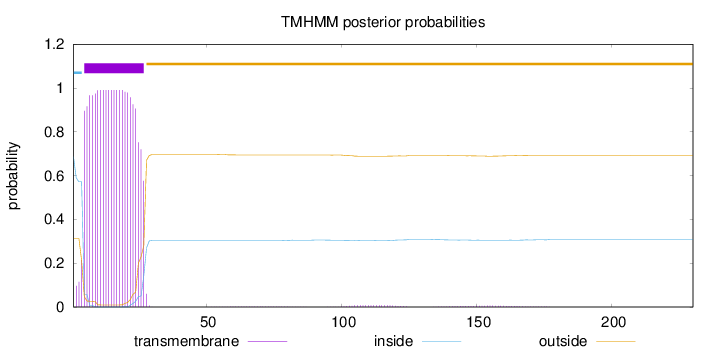
Length:
230
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.2768
Exp number, first 60 AAs:
21.92079
Total prob of N-in:
0.68667
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 230
Population Genetic Test Statistics
Pi
293.014068
Theta
39.027322
Tajima's D
1.869836
CLR
0.319551
CSRT
0.859457027148643
Interpretation
Uncertain
