Pre Gene Modal
BGIBMGA010467
Annotation
PREDICTED:_bifunctional_ATP-dependent_dihydroxyacetone_kinase/FAD-AMP_lyase_(cyclizing)-like?_partial_[Bombyx_mori]
Full name
Triokinase/FMN cyclase
Alternative Name
Bifunctional ATP-dependent dihydroxyacetone kinase/FAD-AMP lyase (cyclizing)
Location in the cell
Cytoplasmic Reliability : 2.362
Sequence
CDS
ATGCTTGGCTTTCAGGATGACAGTAAGAGAGTGGCCATCCTTGGTGGTGGAGGATCGGGACACGAACCCTTTGCATCAGGATTCATAGGTTCGGGAATGCTCGATGGCGCAGTGGCTGGTGGGGTTTTCGCTTCACCACCTACTGGACATGTATTGTACGGGATAGCGCAACTACACAAATATAATTCAGGGGGAGTTCTTGTTATCATTGGTAATTACACAGGCGATCGGCTAAACTTCGGCAAGGCCATCGAGAAAGCTAAAATAGCAGGAATTAAGGTGGAAGGTCTTATCGTGGGAGAGGATGTGGCTTCAAGCAAGAACAAAACAGGCGGGCGCAGTATGGTCGGAGAAGTACTATTTTATAAGCTATGTGGAGCAATGGCCTTAAAAGGTTACGACCTTGAGACGATACGAAAGACTGCAGTTGAAGCTAACAAAAACATGGCCACCCTAGGCGTGTGCCTCAGCGCTTGCTCTCTGCCAGGTCAGCCGCCGCTGTTCGAGATATCACCTGACGAGATGGAACTCGGCGCTGGTGTACACGGCGAAGCTGGTATAGCGAAAGTCAAAATGGGTACCGCCAAGGAAATCGTCAAGATGCTTCTAGCTATAATTGTTGGTCATTTAAAATTATCACGGGGAGATCGGGTAGCGGTCACAGTGGACAACTTAGGTGGTACTTCGTTCTTGGAAATGAATATCATCAGCGCTGAAACTAAGGATTACTTAGAAAACCACGATATCACGGTAGAACGTGTATACGCGGGGCACTTAAAGACCTCACTAGAGATGCACGGATTTCAAATATGTCTTCTGCTTCTAAACAAGGAGCATGGCGATCTCTTCCTGGAGTTATTGGACGCACCCACTTCAGCTGCAGCTTGGACTGGTGGTGAAATATCCGTGAGGAGTGACGGAGGACATGTCGACGATGAGAGCTTGTTGAAAGGTGACGTTAGGAAGGCGGCTTCGGGCCCGTCTTTGTCAGCGAAACAGCAAGATCTTCTCCGCGAGAGTTTGACCTCCGCTGCAGAAGTACTGATTAAGAGCGAGGCTTTACTGAATAAACTAGACTCAGGGTGCGGAGATGGCGACTGCGGGAATACGCTTAAAAAGTTTGGACACGCCATCTTAGCATATCTGAAGACAGCTTCGTTAGAATATGTGTCCAACGTACTCTGGGACTTGTCAGAAATCGCCGAGACCGACATGGGCGGGACCTCGGGTGGAATCTACAGCTTGGGGCTTTCGGCTGCATCACAGGCATTTTCAATAGCTACGTCCAACTCTGTCCCAACATGGCTGTCGGCGTTGGAGAGCGCAATACAAGCTATTTCAAAATACGGCGGTGCTGAGCCCGGCGATAGGACAATGTTGGACACTTTGCATGCAGTAGCTGAAACGTTGAAGAAGTGTATCGAAGAAGAACTGTTAGCGGTGCTGAAGAAGGCTGCAGAAGCTGCCGATCACGGCGCCAAGGCCACAGCGAGTATGGTTGCAAGAGCTGGTAGAGCATCGTACGTGGCTAGCGGTTATACGCAGCAAGAAGACGCTGGGGCAAGAGCAGCAGCTCTTTGGCTACAAGCTATACTGTGTTCTATCGCTAGTAAAATATGA
Protein
MLGFQDDSKRVAILGGGGSGHEPFASGFIGSGMLDGAVAGGVFASPPTGHVLYGIAQLHKYNSGGVLVIIGNYTGDRLNFGKAIEKAKIAGIKVEGLIVGEDVASSKNKTGGRSMVGEVLFYKLCGAMALKGYDLETIRKTAVEANKNMATLGVCLSACSLPGQPPLFEISPDEMELGAGVHGEAGIAKVKMGTAKEIVKMLLAIIVGHLKLSRGDRVAVTVDNLGGTSFLEMNIISAETKDYLENHDITVERVYAGHLKTSLEMHGFQICLLLLNKEHGDLFLELLDAPTSAAAWTGGEISVRSDGGHVDDESLLKGDVRKAASGPSLSAKQQDLLRESLTSAAEVLIKSEALLNKLDSGCGDGDCGNTLKKFGHAILAYLKTASLEYVSNVLWDLSEIAETDMGGTSGGIYSLGLSAASQAFSIATSNSVPTWLSALESAIQAISKYGGAEPGDRTMLDTLHAVAETLKKCIEEELLAVLKKAAEAADHGAKATASMVARAGRASYVASGYTQQEDAGARAAALWLQAILCSIASKI
Summary
Description
Catalyzes both the phosphorylation of dihydroxyacetone and of glyceraldehyde, and the splitting of ribonucleoside diphosphate-X compounds among which FAD is the best substrate. Represses IFIH1-mediated cellular antiviral response (PubMed:17600090).
Catalytic Activity
ATP + dihydroxyacetone = ADP + dihydroxyacetone phosphate + H(+)
ATP + D-glyceraldehyde = ADP + D-glyceraldehyde 3-phosphate + H(+)
FAD = AMP + H(+) + riboflavin cyclic-4',5'-phosphate
ATP + D-glyceraldehyde = ADP + D-glyceraldehyde 3-phosphate + H(+)
FAD = AMP + H(+) + riboflavin cyclic-4',5'-phosphate
Cofactor
Mg(2+)
Biophysicochemical Properties
0.5 uM for dihydroxyacetone
11 uM for glyceraldehyde
1.55 uM for dihydroxyacetone
43.2 uM for ATP
18.1 uM for glyceraldehyde
7 uM for FAD
12 uM for ADP-glucose
317 uM for UDP-glucose
263 uM for UDP-galactose
11 uM for glyceraldehyde
1.55 uM for dihydroxyacetone
43.2 uM for ATP
18.1 uM for glyceraldehyde
7 uM for FAD
12 uM for ADP-glucose
317 uM for UDP-glucose
263 uM for UDP-galactose
Subunit
Homodimer (By similarity). Interacts with IFIH1 (via the CARD domains), the interaction is inhibited by viral infection (PubMed:17600090).
Similarity
Belongs to the dihydroxyacetone kinase (DAK) family.
Keywords
Alternative splicing
ATP-binding
Cobalt
Complete proteome
FAD
Flavoprotein
Kinase
Lyase
Magnesium
Manganese
Metal-binding
Multifunctional enzyme
Nucleotide-binding
Phosphoprotein
Polymorphism
Reference proteome
Transferase
Feature
chain Triokinase/FMN cyclase
splice variant In isoform 2.
sequence variant In dbSNP:rs2260655.
splice variant In isoform 2.
sequence variant In dbSNP:rs2260655.
Uniprot
A0A2A4K9H2
A0A2W1B4U0
A0A194QMW4
A0A194PPA9
A0A2H1VBP3
A0A0L7L3Z9
+ More
A0A212F8X5 A0A154PGQ1 A0A2R2MMQ2 C3ZQZ9 A0A2R2MMP3 A0A1S3IBP5 A0A151INV0 A0A151JT26 F4W7H7 A0A067RGB2 A0A2J7RBU9 A0A026WU04 K1PIX8 A0A1B6D265 A0A2J7RBX7 A0A232ERT0 E2A2U3 A0A088AEP8 A0A2A3EBN0 A0A151X8J8 E9IX13 K7ITH9 E2B9B1 K1PR20 A0A0C9RHJ1 H2XLN7 A0A1W2WHJ9 F1Q8Q6 A0A0H5SDC7 A0A3M6THH3 A0A0B2VND3 A0A1A8NNV3 A0A1A8S4J7 A0A1A8UW57 L8Y5J3 A0A1A8F8N5 A0A3B4CTM6 I3MA42 A0A1A8IZJ0 A0A195AT93 W5MTB7 G1SFW1 A0A158NPY8 C3XZK9 H3BDK7 A0A1S3G900 A0A2B4T0J0 A0A0N4TLW6 Q08CB4 A0A3N0YI63 A0A3Q1AHF4 A0A024R529 W5KUR9 A0A2K5PUJ5 H2ND85 G3QN84 G2HF99 A0A140VJH7 Q3LXA3 F7GX62 H9EV13 A0A2K5KEP8 A0A238C3Z8 A0A2K6KTG4 A0A2K6S7W7 W5ULD7 A0A2D0RAI8 A0A2K6EG49 G1QPV1 D2HUE6 A0A2K6S7X4 F6TNX4 A0A2K6EG62 H3BDK8 A0A096MPA6 A0A1A7XB45 A0A0D9R252 G1MDS9 A0A1A7YB51 A0A2K6BKC7 A0A2K5V408 I0FMH5 G7PPY4 A0A2K5P3L9 A0A2D0RCG5 U3KBU5 A0A2D0RBR1 A0A2K6P8B6 A0A2K5EX23 A0A2U3XGC2 A0A212DGA4 A0A3Q7PM29 T1IL93
A0A212F8X5 A0A154PGQ1 A0A2R2MMQ2 C3ZQZ9 A0A2R2MMP3 A0A1S3IBP5 A0A151INV0 A0A151JT26 F4W7H7 A0A067RGB2 A0A2J7RBU9 A0A026WU04 K1PIX8 A0A1B6D265 A0A2J7RBX7 A0A232ERT0 E2A2U3 A0A088AEP8 A0A2A3EBN0 A0A151X8J8 E9IX13 K7ITH9 E2B9B1 K1PR20 A0A0C9RHJ1 H2XLN7 A0A1W2WHJ9 F1Q8Q6 A0A0H5SDC7 A0A3M6THH3 A0A0B2VND3 A0A1A8NNV3 A0A1A8S4J7 A0A1A8UW57 L8Y5J3 A0A1A8F8N5 A0A3B4CTM6 I3MA42 A0A1A8IZJ0 A0A195AT93 W5MTB7 G1SFW1 A0A158NPY8 C3XZK9 H3BDK7 A0A1S3G900 A0A2B4T0J0 A0A0N4TLW6 Q08CB4 A0A3N0YI63 A0A3Q1AHF4 A0A024R529 W5KUR9 A0A2K5PUJ5 H2ND85 G3QN84 G2HF99 A0A140VJH7 Q3LXA3 F7GX62 H9EV13 A0A2K5KEP8 A0A238C3Z8 A0A2K6KTG4 A0A2K6S7W7 W5ULD7 A0A2D0RAI8 A0A2K6EG49 G1QPV1 D2HUE6 A0A2K6S7X4 F6TNX4 A0A2K6EG62 H3BDK8 A0A096MPA6 A0A1A7XB45 A0A0D9R252 G1MDS9 A0A1A7YB51 A0A2K6BKC7 A0A2K5V408 I0FMH5 G7PPY4 A0A2K5P3L9 A0A2D0RCG5 U3KBU5 A0A2D0RBR1 A0A2K6P8B6 A0A2K5EX23 A0A2U3XGC2 A0A212DGA4 A0A3Q7PM29 T1IL93
Pubmed
28756777
26354079
26227816
22118469
26383154
18563158
+ More
21719571 24845553 24508170 22992520 28648823 20798317 21282665 20075255 12481130 15114417 23594743 17885136 30382153 23385571 21993624 21347285 9215903 11181995 25329095 22398555 16136131 21484476 16289032 14702039 16554811 15489334 4688871 17600090 21269460 24569995 24275569 25319552 23127152 20010809 19892987 17431167 22002653 25362486 29294181
21719571 24845553 24508170 22992520 28648823 20798317 21282665 20075255 12481130 15114417 23594743 17885136 30382153 23385571 21993624 21347285 9215903 11181995 25329095 22398555 16136131 21484476 16289032 14702039 16554811 15489334 4688871 17600090 21269460 24569995 24275569 25319552 23127152 20010809 19892987 17431167 22002653 25362486 29294181
EMBL
NWSH01000039
PCG80380.1
KZ150341
PZC71309.1
KQ461194
KPJ06868.1
+ More
KQ459603 KPI92970.1 ODYU01001690 SOQ38259.1 JTDY01003081 KOB70182.1 AGBW02009660 OWR50180.1 KQ434893 KZC10644.1 GG666663 EEN45040.1 KQ976914 KYN07115.1 KQ982014 KYN30670.1 GL887844 EGI69850.1 KK852482 KDR22916.1 NEVH01005889 PNF38316.1 KK107105 EZA59458.1 JH817601 EKC18869.1 GEDC01017507 JAS19791.1 PNF38318.1 NNAY01002582 OXU20996.1 GL436214 EFN72250.1 KZ288301 PBC28874.1 KQ982409 KYQ56693.1 GL766616 EFZ14941.1 AAZX01003331 GL446479 EFN87717.1 EKC18870.1 GBYB01006396 JAG76163.1 EAAA01001926 BX957278 LN855247 CRZ21959.1 RCHS01003571 RMX40845.1 JPKZ01001291 KHN82859.1 HAEG01004056 SBR70671.1 HAEH01021967 SBS13171.1 HAEJ01011369 SBS51826.1 KB368149 ELV10250.1 HAEB01008599 HAEC01011888 SBQ55126.1 AGTP01066789 HAED01016276 SBR02721.1 KQ976745 KYM75443.1 AHAT01015510 AHAT01015511 AHAT01015512 ADTU01022711 ADTU01022712 GG666475 EEN66530.1 AFYH01010944 AFYH01010945 AFYH01010946 AFYH01010947 LSMT01000001 PFX34853.1 UZAD01013153 VDN90550.1 BC124306 AAI24307.1 RJVU01042551 ROL45561.1 CH471076 EAW73940.1 ABGA01076013 NDHI03003558 PNJ22124.1 CABD030079452 AACZ04057297 AK305413 GABC01002456 GABF01000624 GABD01010396 GABE01006961 NBAG03000297 BAK62407.1 JAA08882.1 JAA21521.1 JAA22704.1 JAA37778.1 PNI45179.1 HM005364 AEE60964.1 DQ138290 DQ344550 AK023915 AK223580 AP003108 BC001341 JU322466 AFE66222.1 KZ269978 OZC12192.1 JT419119 AHH42964.1 ADFV01078994 GL193398 EFB20941.1 AHZZ02006390 HADW01013700 SBP15100.1 AQIB01079262 ACTA01020162 HADX01005132 SBP27364.1 AQIA01020060 JSUE03012184 JV045580 JV636090 AFI35651.1 AFJ71430.1 CM001289 EHH56174.1 AGTO01011732 MKHE01000002 OWK17273.1 JH430755
KQ459603 KPI92970.1 ODYU01001690 SOQ38259.1 JTDY01003081 KOB70182.1 AGBW02009660 OWR50180.1 KQ434893 KZC10644.1 GG666663 EEN45040.1 KQ976914 KYN07115.1 KQ982014 KYN30670.1 GL887844 EGI69850.1 KK852482 KDR22916.1 NEVH01005889 PNF38316.1 KK107105 EZA59458.1 JH817601 EKC18869.1 GEDC01017507 JAS19791.1 PNF38318.1 NNAY01002582 OXU20996.1 GL436214 EFN72250.1 KZ288301 PBC28874.1 KQ982409 KYQ56693.1 GL766616 EFZ14941.1 AAZX01003331 GL446479 EFN87717.1 EKC18870.1 GBYB01006396 JAG76163.1 EAAA01001926 BX957278 LN855247 CRZ21959.1 RCHS01003571 RMX40845.1 JPKZ01001291 KHN82859.1 HAEG01004056 SBR70671.1 HAEH01021967 SBS13171.1 HAEJ01011369 SBS51826.1 KB368149 ELV10250.1 HAEB01008599 HAEC01011888 SBQ55126.1 AGTP01066789 HAED01016276 SBR02721.1 KQ976745 KYM75443.1 AHAT01015510 AHAT01015511 AHAT01015512 ADTU01022711 ADTU01022712 GG666475 EEN66530.1 AFYH01010944 AFYH01010945 AFYH01010946 AFYH01010947 LSMT01000001 PFX34853.1 UZAD01013153 VDN90550.1 BC124306 AAI24307.1 RJVU01042551 ROL45561.1 CH471076 EAW73940.1 ABGA01076013 NDHI03003558 PNJ22124.1 CABD030079452 AACZ04057297 AK305413 GABC01002456 GABF01000624 GABD01010396 GABE01006961 NBAG03000297 BAK62407.1 JAA08882.1 JAA21521.1 JAA22704.1 JAA37778.1 PNI45179.1 HM005364 AEE60964.1 DQ138290 DQ344550 AK023915 AK223580 AP003108 BC001341 JU322466 AFE66222.1 KZ269978 OZC12192.1 JT419119 AHH42964.1 ADFV01078994 GL193398 EFB20941.1 AHZZ02006390 HADW01013700 SBP15100.1 AQIB01079262 ACTA01020162 HADX01005132 SBP27364.1 AQIA01020060 JSUE03012184 JV045580 JV636090 AFI35651.1 AFJ71430.1 CM001289 EHH56174.1 AGTO01011732 MKHE01000002 OWK17273.1 JH430755
Proteomes
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
UP000076502
+ More
UP000085678 UP000001554 UP000078542 UP000078541 UP000007755 UP000027135 UP000235965 UP000053097 UP000005408 UP000215335 UP000000311 UP000005203 UP000242457 UP000075809 UP000002358 UP000008237 UP000008144 UP000000437 UP000006672 UP000275408 UP000031036 UP000011518 UP000261440 UP000005215 UP000078540 UP000018468 UP000001811 UP000005205 UP000008672 UP000081671 UP000225706 UP000038020 UP000278627 UP000257160 UP000018467 UP000233040 UP000001595 UP000001519 UP000002277 UP000005640 UP000008225 UP000233080 UP000233180 UP000233220 UP000221080 UP000233160 UP000001073 UP000002281 UP000028761 UP000029965 UP000008912 UP000233120 UP000233100 UP000006718 UP000009130 UP000233060 UP000016665 UP000233200 UP000233020 UP000245341 UP000286641
UP000085678 UP000001554 UP000078542 UP000078541 UP000007755 UP000027135 UP000235965 UP000053097 UP000005408 UP000215335 UP000000311 UP000005203 UP000242457 UP000075809 UP000002358 UP000008237 UP000008144 UP000000437 UP000006672 UP000275408 UP000031036 UP000011518 UP000261440 UP000005215 UP000078540 UP000018468 UP000001811 UP000005205 UP000008672 UP000081671 UP000225706 UP000038020 UP000278627 UP000257160 UP000018467 UP000233040 UP000001595 UP000001519 UP000002277 UP000005640 UP000008225 UP000233080 UP000233180 UP000233220 UP000221080 UP000233160 UP000001073 UP000002281 UP000028761 UP000029965 UP000008912 UP000233120 UP000233100 UP000006718 UP000009130 UP000233060 UP000016665 UP000233200 UP000233020 UP000245341 UP000286641
PRIDE
SUPFAM
SSF101473
SSF101473
Gene 3D
ProteinModelPortal
A0A2A4K9H2
A0A2W1B4U0
A0A194QMW4
A0A194PPA9
A0A2H1VBP3
A0A0L7L3Z9
+ More
A0A212F8X5 A0A154PGQ1 A0A2R2MMQ2 C3ZQZ9 A0A2R2MMP3 A0A1S3IBP5 A0A151INV0 A0A151JT26 F4W7H7 A0A067RGB2 A0A2J7RBU9 A0A026WU04 K1PIX8 A0A1B6D265 A0A2J7RBX7 A0A232ERT0 E2A2U3 A0A088AEP8 A0A2A3EBN0 A0A151X8J8 E9IX13 K7ITH9 E2B9B1 K1PR20 A0A0C9RHJ1 H2XLN7 A0A1W2WHJ9 F1Q8Q6 A0A0H5SDC7 A0A3M6THH3 A0A0B2VND3 A0A1A8NNV3 A0A1A8S4J7 A0A1A8UW57 L8Y5J3 A0A1A8F8N5 A0A3B4CTM6 I3MA42 A0A1A8IZJ0 A0A195AT93 W5MTB7 G1SFW1 A0A158NPY8 C3XZK9 H3BDK7 A0A1S3G900 A0A2B4T0J0 A0A0N4TLW6 Q08CB4 A0A3N0YI63 A0A3Q1AHF4 A0A024R529 W5KUR9 A0A2K5PUJ5 H2ND85 G3QN84 G2HF99 A0A140VJH7 Q3LXA3 F7GX62 H9EV13 A0A2K5KEP8 A0A238C3Z8 A0A2K6KTG4 A0A2K6S7W7 W5ULD7 A0A2D0RAI8 A0A2K6EG49 G1QPV1 D2HUE6 A0A2K6S7X4 F6TNX4 A0A2K6EG62 H3BDK8 A0A096MPA6 A0A1A7XB45 A0A0D9R252 G1MDS9 A0A1A7YB51 A0A2K6BKC7 A0A2K5V408 I0FMH5 G7PPY4 A0A2K5P3L9 A0A2D0RCG5 U3KBU5 A0A2D0RBR1 A0A2K6P8B6 A0A2K5EX23 A0A2U3XGC2 A0A212DGA4 A0A3Q7PM29 T1IL93
A0A212F8X5 A0A154PGQ1 A0A2R2MMQ2 C3ZQZ9 A0A2R2MMP3 A0A1S3IBP5 A0A151INV0 A0A151JT26 F4W7H7 A0A067RGB2 A0A2J7RBU9 A0A026WU04 K1PIX8 A0A1B6D265 A0A2J7RBX7 A0A232ERT0 E2A2U3 A0A088AEP8 A0A2A3EBN0 A0A151X8J8 E9IX13 K7ITH9 E2B9B1 K1PR20 A0A0C9RHJ1 H2XLN7 A0A1W2WHJ9 F1Q8Q6 A0A0H5SDC7 A0A3M6THH3 A0A0B2VND3 A0A1A8NNV3 A0A1A8S4J7 A0A1A8UW57 L8Y5J3 A0A1A8F8N5 A0A3B4CTM6 I3MA42 A0A1A8IZJ0 A0A195AT93 W5MTB7 G1SFW1 A0A158NPY8 C3XZK9 H3BDK7 A0A1S3G900 A0A2B4T0J0 A0A0N4TLW6 Q08CB4 A0A3N0YI63 A0A3Q1AHF4 A0A024R529 W5KUR9 A0A2K5PUJ5 H2ND85 G3QN84 G2HF99 A0A140VJH7 Q3LXA3 F7GX62 H9EV13 A0A2K5KEP8 A0A238C3Z8 A0A2K6KTG4 A0A2K6S7W7 W5ULD7 A0A2D0RAI8 A0A2K6EG49 G1QPV1 D2HUE6 A0A2K6S7X4 F6TNX4 A0A2K6EG62 H3BDK8 A0A096MPA6 A0A1A7XB45 A0A0D9R252 G1MDS9 A0A1A7YB51 A0A2K6BKC7 A0A2K5V408 I0FMH5 G7PPY4 A0A2K5P3L9 A0A2D0RCG5 U3KBU5 A0A2D0RBR1 A0A2K6P8B6 A0A2K5EX23 A0A2U3XGC2 A0A212DGA4 A0A3Q7PM29 T1IL93
PDB
1UN8
E-value=1.1933e-47,
Score=480
Ontologies
PATHWAY
GO
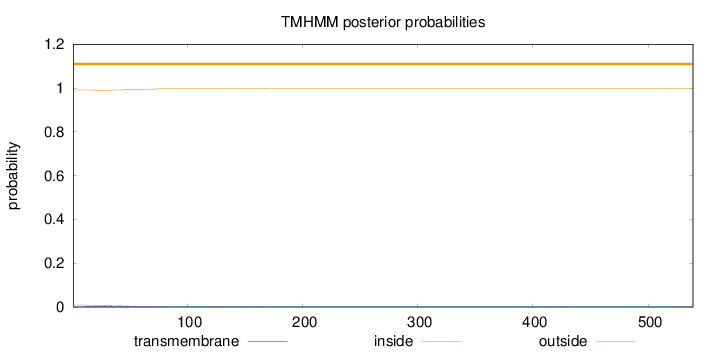
Topology
Length:
539
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29933
Exp number, first 60 AAs:
0.23288
Total prob of N-in:
0.00942
outside
1 - 539
Population Genetic Test Statistics
Pi
201.494354
Theta
149.017681
Tajima's D
0.818716
CLR
0.174285
CSRT
0.599570021498925
Interpretation
Uncertain
