Pre Gene Modal
BGIBMGA010473
Annotation
Dynein-1-beta_heavy_chain?_flagellar_inner_arm_I1_complex_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.401 Nuclear Reliability : 1.222 PlasmaMembrane Reliability : 1.156
Sequence
CDS
ATGCTCGGTCTCACCGTATTGTATATACCTAGAGAGGGACAGCAGCTGACGTTCGAACAGGCCAGCGCAGACCGGGAGCTGGTGAAAAGGCTGGAAGGGGTTGTGGTATACTGGACGCATCAGATCAAATCGTGCTTGGAGGACCATGCGTTTGTGGCGTCACAGAAAGAGCTCTTGTGCCCGAGTGATGAATACGACTTTTGGATATATCGGCCACCGACCGTTTCAGACGAAAACTTAAATGCCCTGCTATACCAATTGAAGAATCCAGCGGTAAGACACATCACGAAAACACTGGTGACCACACACTCGACTTTCATTCACCAATTCCAAACTCTTTGCCAAGAAGTCATTCAGAAGATCAACGAAGCCACTTCCAACATTGAATACTTACAAGTGATCAAACAGCCTTGTGCGATTTTGGAATGCGTCATAGATCCCGATGAAATAGCCAAACACGTTCCCCAAATAATCAATCTGTTCAGATTTATCTGGATGGAGTCCCCATATTACAACTCCGAGGCCAGAATAACAAATTTATTCAAGGCCCTGAGCAATCAAATCATAATACTGTGCAGGAATTACGTAAATCTAGACGAGCTGTTTGAGGGGGAAACTAAGAAAGCGCTCGGTGAATTCACAAAGTGCATTGAGTGTTGCAAGAAATACCGCGAAATTTACGACACGATGGCCGAGGCTCACTGCGATTCGAAGCCAGACTCTTGGAATTTGGACACTGGTAGCATTTTCAATTACATCGATTCCTTCGTTCAGAGGTGCTTTGATATGTTAGAAGTTTGTAACTGCATGATTATTTTTGGAAGGATCGACGAACTAGAACACATCAATAAGCCGATGTTCAGTGGCGCTCGCGGTGACCAGTTCGAGGCTAAATGCGATCAAATTGAGCACATGTTTCAAGATGCGCTACTCAACGTCCGCAGAGTGTCGCGCGCCATACTAGACGTCCAGGCGCCGGCCTGGTACGACGATATCCTGCAGTTCAGGACTGTCATCAAGGATATTGAGATTATCATAGAGAACTTGGTGGAGTCAGTCTTTGAAGGTGTGAATCACGTGGAAGAGGCTGTGGTCGCTCTGTTTTCGCTCAACAATTACTCCAAGCGGAAGAGTTTGAAACGGATTTTCAAGAAGAAAACTGCAGAGGTTTGGGCGATGTTTAGCGACGAGGTACAGGAAGCTAAAAAGGATATGGTCTCGAATCGAGGTCAGTTTCCAGCTGATCTGCCGTCCTTTGCAGGGCGAGCTATTGTACTCAAGACGAGGAAAAACAGACTTGTGTACTTGAAAAAGGTGATGACTGATGCCTCAGTATGGATGATGGCGTGCAGTAACTCGGAAGACGTTATCATGCACGTAAACAGACTGATGGGCGCCATCGATGTTGCCATACGCGAACTGTGGATTTCCTGGACGCACAATTTAGATGAAAAATGTAGCGCGGGCTTGAACAGGACCTTGATGCGGAAGAGCGCAGAGAACTTAGGACTCTTGGAGTGCAACATCGATATGAACATTTTGGATCTTTGTAAGGAGGCAGCACATTGGGAAAACTTGGGACAAGATATCCCATTGCACGCTTATCAGGTATACATGAAAAGCAAAACTATTTACTATGTTTACGAAAGCGTGTTGGCCGTGGTTAAGGGATACAACAAGATAATCGACTCGCTGTCTGACGATGAAAGATTACTCTTCAAGCCTATGATTACAGTGAGTATATACTTTCATCCATTGGCTGTTTATTATAAAGACTGA
Protein
MLGLTVLYIPREGQQLTFEQASADRELVKRLEGVVVYWTHQIKSCLEDHAFVASQKELLCPSDEYDFWIYRPPTVSDENLNALLYQLKNPAVRHITKTLVTTHSTFIHQFQTLCQEVIQKINEATSNIEYLQVIKQPCAILECVIDPDEIAKHVPQIINLFRFIWMESPYYNSEARITNLFKALSNQIIILCRNYVNLDELFEGETKKALGEFTKCIECCKKYREIYDTMAEAHCDSKPDSWNLDTGSIFNYIDSFVQRCFDMLEVCNCMIIFGRIDELEHINKPMFSGARGDQFEAKCDQIEHMFQDALLNVRRVSRAILDVQAPAWYDDILQFRTVIKDIEIIIENLVESVFEGVNHVEEAVVALFSLNNYSKRKSLKRIFKKKTAEVWAMFSDEVQEAKKDMVSNRGQFPADLPSFAGRAIVLKTRKNRLVYLKKVMTDASVWMMACSNSEDVIMHVNRLMGAIDVAIRELWISWTHNLDEKCSAGLNRTLMRKSAENLGLLECNIDMNILDLCKEAAHWENLGQDIPLHAYQVYMKSKTIYYVYESVLAVVKGYNKIIDSLSDDERLLFKPMITVSIYFHPLAVYYKD
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9JLS1
A0A194PIV2
A0A194QNC6
A0A2A4K516
A0A3S2LVM1
A0A2W1BIP9
+ More
A0A2H1WSD3 A0A212F756 A0A0L7LKZ3 U4U449 N6TGV9 A0A139WBF9 A0A0J7KPM0 A0A154PKQ9 A0A026VYL0 A0A195FFT1 E2C2D9 A0A0L7RJF9 A0A151J3Q5 E1ZVJ0 A0A151WED7 A0A151IDR4 A0A158NBM9 E0VRS1 F4WJJ9 A0A310SN52 A0A3L8E1T2 A0A088AGE0 A0A232FJ76 K7J522 A0A034VMG8 A0A0A1XRG0 A0A0R3P0Q9 A0A0L0CHS0 B4YLL8 A0A1I8MW68 A0A0K8W9W9 A0A1B6CE00 A0A0R3NW00 A0A1A9ZES5 A0A1B0C1D9 B4YLL9 A0A1B0C1D8 A0A1B0A5J4 B4YLL6 B4YLL7 A0A1A9X8E5 A0A1A9X8E6 A0A1B0G1Q7 C3ZEM0 A0A1B6FLY6 K1PG48 A0A1S3SDP5 A0A1W5BAL8 A0A210QEN0 W4Y9K3 A0A1J1I591 A0A1I8PQ03 A0A0U1RVI8 Q5LJP0 A0A182R127 A0A369SGH2 A0A3M7S4K3 W4ZLH2 E4YLH2 A0A182TF21 B4YLM5 B4YLM3 A0A2Y9D2Y0 A0A182JPP7 A0A1S4H272 A0A2Y9D1B9 A0A2Y9D3H5 A0A1W4VSS7 B4MZY1 H2ZI78 A0A1D5NT12 A0A084VP90 A0A3M6T609 A0A182Y5Q8 A0A1L8H3M0 A0A1Y9GLY7 A0A1I8J9I4 A0A1I8H908 A0A1I8GHU8 A0A1I8J8T8 A0A182PB45 A0A182RPQ4 H2ZI79 W5J4X2 A0A0F8AII0 A0A182F4F5 B4HSV3 W5N7B2 A0A182M2Z6 A0A0J9RE18 W5N7C9 A1ZAD3 A0A182VV30 A0A182NEB2
A0A2H1WSD3 A0A212F756 A0A0L7LKZ3 U4U449 N6TGV9 A0A139WBF9 A0A0J7KPM0 A0A154PKQ9 A0A026VYL0 A0A195FFT1 E2C2D9 A0A0L7RJF9 A0A151J3Q5 E1ZVJ0 A0A151WED7 A0A151IDR4 A0A158NBM9 E0VRS1 F4WJJ9 A0A310SN52 A0A3L8E1T2 A0A088AGE0 A0A232FJ76 K7J522 A0A034VMG8 A0A0A1XRG0 A0A0R3P0Q9 A0A0L0CHS0 B4YLL8 A0A1I8MW68 A0A0K8W9W9 A0A1B6CE00 A0A0R3NW00 A0A1A9ZES5 A0A1B0C1D9 B4YLL9 A0A1B0C1D8 A0A1B0A5J4 B4YLL6 B4YLL7 A0A1A9X8E5 A0A1A9X8E6 A0A1B0G1Q7 C3ZEM0 A0A1B6FLY6 K1PG48 A0A1S3SDP5 A0A1W5BAL8 A0A210QEN0 W4Y9K3 A0A1J1I591 A0A1I8PQ03 A0A0U1RVI8 Q5LJP0 A0A182R127 A0A369SGH2 A0A3M7S4K3 W4ZLH2 E4YLH2 A0A182TF21 B4YLM5 B4YLM3 A0A2Y9D2Y0 A0A182JPP7 A0A1S4H272 A0A2Y9D1B9 A0A2Y9D3H5 A0A1W4VSS7 B4MZY1 H2ZI78 A0A1D5NT12 A0A084VP90 A0A3M6T609 A0A182Y5Q8 A0A1L8H3M0 A0A1Y9GLY7 A0A1I8J9I4 A0A1I8H908 A0A1I8GHU8 A0A1I8J8T8 A0A182PB45 A0A182RPQ4 H2ZI79 W5J4X2 A0A0F8AII0 A0A182F4F5 B4HSV3 W5N7B2 A0A182M2Z6 A0A0J9RE18 W5N7C9 A1ZAD3 A0A182VV30 A0A182NEB2
Pubmed
19121390
26354079
28756777
22118469
26227816
23537049
+ More
18362917 19820115 24508170 20798317 21347285 20566863 21719571 30249741 28648823 20075255 25348373 25830018 15632085 17994087 26108605 25315136 18563158 22992520 28812685 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30042472 30375419 21097902 12364791 20966253 23594743 24438588 30382153 25244985 27762356 26392545 20920257 23761445 25835551 22936249
18362917 19820115 24508170 20798317 21347285 20566863 21719571 30249741 28648823 20075255 25348373 25830018 15632085 17994087 26108605 25315136 18563158 22992520 28812685 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30042472 30375419 21097902 12364791 20966253 23594743 24438588 30382153 25244985 27762356 26392545 20920257 23761445 25835551 22936249
EMBL
BABH01005183
KQ459603
KPI92978.1
KQ461194
KPJ06859.1
NWSH01000120
+ More
PCG79345.1 RSAL01000003 RVE54736.1 KZ150161 PZC72706.1 ODYU01010314 SOQ55334.1 AGBW02009920 OWR49571.1 JTDY01000688 KOB76208.1 KB631679 ERL85346.1 APGK01038540 KB740960 ENN77013.1 KQ971372 KYB25181.1 LBMM01004643 KMQ92211.1 KQ434948 KZC12413.1 KK107574 EZA48745.1 KQ981606 KYN39560.1 GL452124 EFN77889.1 KQ414581 KOC70969.1 KQ980259 KYN17075.1 GL434547 EFN74800.1 KQ983238 KYQ46233.1 KQ977978 KYM98389.1 ADTU01000355 ADTU01000356 ADTU01000357 ADTU01000358 ADTU01000359 DS235494 EEB16077.1 GL888184 EGI65615.1 KQ759821 OAD62685.1 QOIP01000002 RLU26149.1 NNAY01000118 OXU30796.1 AAZX01000330 GAKP01015942 JAC43010.1 GBXI01000730 JAD13562.1 CH475573 KRT05117.1 JRES01000383 KNC31762.1 EU595399 ACC62142.1 GDHF01004437 JAI47877.1 GEDC01025778 JAS11520.1 KRT05116.1 JXJN01023990 EU595400 ACC62143.2 JXJN01023991 EU595397 ACC62140.2 EU595398 ACC62141.2 CCAG010017219 GG666612 EEN49176.1 GECZ01018553 JAS51216.1 JH815791 EKC22887.1 NEDP02004020 OWF47169.1 AAGJ04130935 CVRI01000037 CRK93545.1 EU685283 ACE79738.1 CP007106 EAL24531.5 AXCN02000864 NOWV01000011 RDD46015.1 REGN01002048 RNA30736.1 AAGJ04130936 AAGJ04130937 AAGJ04130938 AAGJ04130939 AAGJ04130940 AAGJ04130941 AAGJ04130942 AAGJ04130943 AAGJ04130944 FN654759 CBY36333.1 EU595403 ACC62149.2 EU595402 ACC62147.2 AAAB01008839 CH963920 EDW77916.1 AL935046 AL954772 CU570992 ATLV01014985 KE524999 KFB39784.1 RCHS01004213 RMX36856.1 CM004470 OCT90707.1 APCN01004546 ADMH02002133 ETN58448.1 KQ041833 KKF21926.1 CH480816 EDW48117.1 AHAT01030211 AHAT01030212 AHAT01030213 AHAT01030214 AHAT01030215 AHAT01030216 AHAT01030217 AHAT01030218 AHAT01030219 AHAT01030220 AXCM01000166 CM002911 KMY94232.1 AE013599 AAF58022.1
PCG79345.1 RSAL01000003 RVE54736.1 KZ150161 PZC72706.1 ODYU01010314 SOQ55334.1 AGBW02009920 OWR49571.1 JTDY01000688 KOB76208.1 KB631679 ERL85346.1 APGK01038540 KB740960 ENN77013.1 KQ971372 KYB25181.1 LBMM01004643 KMQ92211.1 KQ434948 KZC12413.1 KK107574 EZA48745.1 KQ981606 KYN39560.1 GL452124 EFN77889.1 KQ414581 KOC70969.1 KQ980259 KYN17075.1 GL434547 EFN74800.1 KQ983238 KYQ46233.1 KQ977978 KYM98389.1 ADTU01000355 ADTU01000356 ADTU01000357 ADTU01000358 ADTU01000359 DS235494 EEB16077.1 GL888184 EGI65615.1 KQ759821 OAD62685.1 QOIP01000002 RLU26149.1 NNAY01000118 OXU30796.1 AAZX01000330 GAKP01015942 JAC43010.1 GBXI01000730 JAD13562.1 CH475573 KRT05117.1 JRES01000383 KNC31762.1 EU595399 ACC62142.1 GDHF01004437 JAI47877.1 GEDC01025778 JAS11520.1 KRT05116.1 JXJN01023990 EU595400 ACC62143.2 JXJN01023991 EU595397 ACC62140.2 EU595398 ACC62141.2 CCAG010017219 GG666612 EEN49176.1 GECZ01018553 JAS51216.1 JH815791 EKC22887.1 NEDP02004020 OWF47169.1 AAGJ04130935 CVRI01000037 CRK93545.1 EU685283 ACE79738.1 CP007106 EAL24531.5 AXCN02000864 NOWV01000011 RDD46015.1 REGN01002048 RNA30736.1 AAGJ04130936 AAGJ04130937 AAGJ04130938 AAGJ04130939 AAGJ04130940 AAGJ04130941 AAGJ04130942 AAGJ04130943 AAGJ04130944 FN654759 CBY36333.1 EU595403 ACC62149.2 EU595402 ACC62147.2 AAAB01008839 CH963920 EDW77916.1 AL935046 AL954772 CU570992 ATLV01014985 KE524999 KFB39784.1 RCHS01004213 RMX36856.1 CM004470 OCT90707.1 APCN01004546 ADMH02002133 ETN58448.1 KQ041833 KKF21926.1 CH480816 EDW48117.1 AHAT01030211 AHAT01030212 AHAT01030213 AHAT01030214 AHAT01030215 AHAT01030216 AHAT01030217 AHAT01030218 AHAT01030219 AHAT01030220 AXCM01000166 CM002911 KMY94232.1 AE013599 AAF58022.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000283053
UP000007151
+ More
UP000037510 UP000030742 UP000019118 UP000007266 UP000036403 UP000076502 UP000053097 UP000078541 UP000008237 UP000053825 UP000078492 UP000000311 UP000075809 UP000078542 UP000005205 UP000009046 UP000007755 UP000279307 UP000005203 UP000215335 UP000002358 UP000001819 UP000037069 UP000095301 UP000092445 UP000092460 UP000092443 UP000092444 UP000001554 UP000005408 UP000087266 UP000242188 UP000007110 UP000183832 UP000095300 UP000000803 UP000075886 UP000253843 UP000276133 UP000075902 UP000075903 UP000075881 UP000075882 UP000076407 UP000192221 UP000007798 UP000007875 UP000000437 UP000030765 UP000275408 UP000076408 UP000186698 UP000075840 UP000095280 UP000075885 UP000075900 UP000000673 UP000069272 UP000001292 UP000018468 UP000075883 UP000075920 UP000075884
UP000037510 UP000030742 UP000019118 UP000007266 UP000036403 UP000076502 UP000053097 UP000078541 UP000008237 UP000053825 UP000078492 UP000000311 UP000075809 UP000078542 UP000005205 UP000009046 UP000007755 UP000279307 UP000005203 UP000215335 UP000002358 UP000001819 UP000037069 UP000095301 UP000092445 UP000092460 UP000092443 UP000092444 UP000001554 UP000005408 UP000087266 UP000242188 UP000007110 UP000183832 UP000095300 UP000000803 UP000075886 UP000253843 UP000276133 UP000075902 UP000075903 UP000075881 UP000075882 UP000076407 UP000192221 UP000007798 UP000007875 UP000000437 UP000030765 UP000275408 UP000076408 UP000186698 UP000075840 UP000095280 UP000075885 UP000075900 UP000000673 UP000069272 UP000001292 UP000018468 UP000075883 UP000075920 UP000075884
Pfam
Interpro
IPR013602
Dynein_heavy_dom-2
+ More
IPR013594 Dynein_heavy_dom-1
IPR042222 Dynein_2_N
IPR042228 Dynein_2_C
IPR024317 Dynein_heavy_chain_D4_dom
IPR035699 AAA_6
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR041589 DNAH3_AAA_lid_1
IPR024743 Dynein_HC_stalk
IPR035706 AAA_9
IPR041658 AAA_lid_11
IPR041228 Dynein_C
IPR004273 Dynein_heavy_D6_P-loop
IPR042219 AAA_lid_11_sf
IPR008952 Tetraspanin_EC2_sf
IPR018499 Tetraspanin/Peripherin
IPR018247 EF_Hand_1_Ca_BS
IPR026983 DHC_fam
IPR000863 Sulfotransferase_dom
IPR013594 Dynein_heavy_dom-1
IPR042222 Dynein_2_N
IPR042228 Dynein_2_C
IPR024317 Dynein_heavy_chain_D4_dom
IPR035699 AAA_6
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR027417 P-loop_NTPase
IPR041589 DNAH3_AAA_lid_1
IPR024743 Dynein_HC_stalk
IPR035706 AAA_9
IPR041658 AAA_lid_11
IPR041228 Dynein_C
IPR004273 Dynein_heavy_D6_P-loop
IPR042219 AAA_lid_11_sf
IPR008952 Tetraspanin_EC2_sf
IPR018499 Tetraspanin/Peripherin
IPR018247 EF_Hand_1_Ca_BS
IPR026983 DHC_fam
IPR000863 Sulfotransferase_dom
ProteinModelPortal
H9JLS1
A0A194PIV2
A0A194QNC6
A0A2A4K516
A0A3S2LVM1
A0A2W1BIP9
+ More
A0A2H1WSD3 A0A212F756 A0A0L7LKZ3 U4U449 N6TGV9 A0A139WBF9 A0A0J7KPM0 A0A154PKQ9 A0A026VYL0 A0A195FFT1 E2C2D9 A0A0L7RJF9 A0A151J3Q5 E1ZVJ0 A0A151WED7 A0A151IDR4 A0A158NBM9 E0VRS1 F4WJJ9 A0A310SN52 A0A3L8E1T2 A0A088AGE0 A0A232FJ76 K7J522 A0A034VMG8 A0A0A1XRG0 A0A0R3P0Q9 A0A0L0CHS0 B4YLL8 A0A1I8MW68 A0A0K8W9W9 A0A1B6CE00 A0A0R3NW00 A0A1A9ZES5 A0A1B0C1D9 B4YLL9 A0A1B0C1D8 A0A1B0A5J4 B4YLL6 B4YLL7 A0A1A9X8E5 A0A1A9X8E6 A0A1B0G1Q7 C3ZEM0 A0A1B6FLY6 K1PG48 A0A1S3SDP5 A0A1W5BAL8 A0A210QEN0 W4Y9K3 A0A1J1I591 A0A1I8PQ03 A0A0U1RVI8 Q5LJP0 A0A182R127 A0A369SGH2 A0A3M7S4K3 W4ZLH2 E4YLH2 A0A182TF21 B4YLM5 B4YLM3 A0A2Y9D2Y0 A0A182JPP7 A0A1S4H272 A0A2Y9D1B9 A0A2Y9D3H5 A0A1W4VSS7 B4MZY1 H2ZI78 A0A1D5NT12 A0A084VP90 A0A3M6T609 A0A182Y5Q8 A0A1L8H3M0 A0A1Y9GLY7 A0A1I8J9I4 A0A1I8H908 A0A1I8GHU8 A0A1I8J8T8 A0A182PB45 A0A182RPQ4 H2ZI79 W5J4X2 A0A0F8AII0 A0A182F4F5 B4HSV3 W5N7B2 A0A182M2Z6 A0A0J9RE18 W5N7C9 A1ZAD3 A0A182VV30 A0A182NEB2
A0A2H1WSD3 A0A212F756 A0A0L7LKZ3 U4U449 N6TGV9 A0A139WBF9 A0A0J7KPM0 A0A154PKQ9 A0A026VYL0 A0A195FFT1 E2C2D9 A0A0L7RJF9 A0A151J3Q5 E1ZVJ0 A0A151WED7 A0A151IDR4 A0A158NBM9 E0VRS1 F4WJJ9 A0A310SN52 A0A3L8E1T2 A0A088AGE0 A0A232FJ76 K7J522 A0A034VMG8 A0A0A1XRG0 A0A0R3P0Q9 A0A0L0CHS0 B4YLL8 A0A1I8MW68 A0A0K8W9W9 A0A1B6CE00 A0A0R3NW00 A0A1A9ZES5 A0A1B0C1D9 B4YLL9 A0A1B0C1D8 A0A1B0A5J4 B4YLL6 B4YLL7 A0A1A9X8E5 A0A1A9X8E6 A0A1B0G1Q7 C3ZEM0 A0A1B6FLY6 K1PG48 A0A1S3SDP5 A0A1W5BAL8 A0A210QEN0 W4Y9K3 A0A1J1I591 A0A1I8PQ03 A0A0U1RVI8 Q5LJP0 A0A182R127 A0A369SGH2 A0A3M7S4K3 W4ZLH2 E4YLH2 A0A182TF21 B4YLM5 B4YLM3 A0A2Y9D2Y0 A0A182JPP7 A0A1S4H272 A0A2Y9D1B9 A0A2Y9D3H5 A0A1W4VSS7 B4MZY1 H2ZI78 A0A1D5NT12 A0A084VP90 A0A3M6T609 A0A182Y5Q8 A0A1L8H3M0 A0A1Y9GLY7 A0A1I8J9I4 A0A1I8H908 A0A1I8GHU8 A0A1I8J8T8 A0A182PB45 A0A182RPQ4 H2ZI79 W5J4X2 A0A0F8AII0 A0A182F4F5 B4HSV3 W5N7B2 A0A182M2Z6 A0A0J9RE18 W5N7C9 A1ZAD3 A0A182VV30 A0A182NEB2
Ontologies
GO
PANTHER
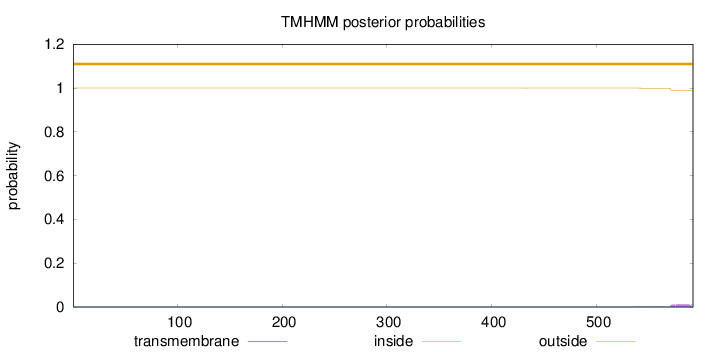
Topology
Length:
592
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00013
outside
1 - 592
Population Genetic Test Statistics
Pi
47.493249
Theta
155.409978
Tajima's D
-1.147176
CLR
133.960668
CSRT
0.113294335283236
Interpretation
Uncertain
