Gene
KWMTBOMO06998 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010476
Annotation
scavenger_receptor_class_B_member_3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.577
Sequence
CDS
ATGAAGATGGTGTCTTCCGGAGTGAAAAGCGGTCTGTTCATGGGCTTCGGGTCAGCGCTGGTCCTTATCGGTGCTATCGTAGTAGTATACTGGCCTTCCCTCTTCATGGCTCAGTTACAGAGGATGATGATCCTATCGCCGACCTCAACGTCGTTCGGTATATGGCAGGAGACTCCAATACCGATGTACCTGGAATGCTACATGTTCAACATAACGAACGCGGATAAGATCCTCGCGAAAGAGGACGTAATACTGAAAGTGGAGCAGTTGGGTCCTTACGTGTTCAGGGAGTCACATTCTAAGGTGAACCTGACTTGGAATGATAACAGCACAATCACATTCTACAATCAGAGATTTTGGCACTTCGACCAGAATTTGTCGAAGGGAAGTCTTTCGGATGAGATTATCAGCATCAACCCTATAATAGCGACTGTGGCGTACATTGTCAGACACCAGCCCAGGGTCGTCAAGGTGTCCGTTGACGTTTTCCTGCGAATGTTCCACGACGACCTCTTCCTGAAGGCGAACGTCTCGTCCTGGCTGTTCGAAGGCATCGAAGACCCGGTTTTGGAGATGGCCCAAAAGTTTCCCGATCTACCCTTGAATATACCGTACGACAAATTCGGATGGTTCTATGAGCGCAATGGCTCGAGGGAATTTGACGGTTCGTTCATAATGAACACGGGAGCGGCGGACTTCTCGCGGCTCGGCAACATTGAGCTGTGGAAGTACTCCCCCAGGACCGTGTTCAGAGACCACTGCGGAGACGTGAAGGGGTCCACCGGGGAGCTATGGGCCCCAGAGCTGGGGCAACCCGAGTTGTTCGTCTTCGCGTCCGACATATGCACATACCTGACACTGCACAAGAAGGAAGACGTGCTGATAGAAAACATAGAGGGCGTTCGGTACGCGGCCAACGACTCGCTGTTCGACAACGGACACAAATATCCCGCAATGGCGTGCTACTGCGACGAGGTTAGGGACCGGGACTGCGTGCCCCCCGGCGCCCTCAACGTGTCCCTGTGCCGCCTGGGAGCCCCGGCCTTCGTGTCCCAGCCGCACTTCCTGGACGCGGACCCGTACTACCCCAGCAAGATCCAGGGTCTGGACCCGCAGGAGGAGCACAGGTTCAGCCTGGCGCTGGAGATGTTCACCGGGATGCCGCTGGCCGTATCCGCGCAGCTGCAGATCAACCTGCTCATCAGAGATGTCTGGGGGATAAGTATCAACAACGTGTTGCCGGACCCGGACACGATGGTGCCGATGTTCTGGTTCCGGCAAGAGCTGCAGGTGACGCCGGAGTACGCGCACATGGCCAGAGTCGCTCTTGCGCTTCGATACTACACTCCATACGCGCTCTACACCCTCACCGTAATCGGCGTGGTCCTCATATTAGTAGGTGTAGTCGTACTAATCAGAAGACTTCTGAATTCATCAGACACGACGCCTCTCATAGAAGAAAGTGCGTCACAAGAGAATCAACAATAA
Protein
MKMVSSGVKSGLFMGFGSALVLIGAIVVVYWPSLFMAQLQRMMILSPTSTSFGIWQETPIPMYLECYMFNITNADKILAKEDVILKVEQLGPYVFRESHSKVNLTWNDNSTITFYNQRFWHFDQNLSKGSLSDEIISINPIIATVAYIVRHQPRVVKVSVDVFLRMFHDDLFLKANVSSWLFEGIEDPVLEMAQKFPDLPLNIPYDKFGWFYERNGSREFDGSFIMNTGAADFSRLGNIELWKYSPRTVFRDHCGDVKGSTGELWAPELGQPELFVFASDICTYLTLHKKEDVLIENIEGVRYAANDSLFDNGHKYPAMACYCDEVRDRDCVPPGALNVSLCRLGAPAFVSQPHFLDADPYYPSKIQGLDPQEEHRFSLALEMFTGMPLAVSAQLQINLLIRDVWGISINNVLPDPDTMVPMFWFRQELQVTPEYAHMARVALALRYYTPYALYTLTVIGVVLILVGVVVLIRRLLNSSDTTPLIEESASQENQQ
Summary
Similarity
Belongs to the CD36 family.
Uniprot
D2KXB2
H9JLS4
A0A1E1WFQ5
A0A1E1WCD1
A0A2A4JQ46
A0A3S2P2C8
+ More
A0A2H1W793 A0A2W1B9R7 A0A194QMT9 A0A194QKR3 A0A212EWB0 A0A212EWD3 A0A0L7RE26 A0A1Y1ME42 A0A310SI06 A0A182K7G5 A0A067RMF2 A0A182TUQ6 A0A0M9A3Q8 A0A182HQX0 A0A182KJJ7 A0A182UZ84 A0A182J4G5 A0A182WXJ4 Q7PMH9 W5JRD6 A0A2M4BM64 A0A2A3EDW6 A0A2M4BLB4 A0A2M4BLB3 A0A2M4BLB8 A0A182QLD0 A0A023ET26 A0A087ZYQ7 E2A0F3 E2C7F5 A0A1W4WU99 A0A1W4WU80 B0W704 A0A1Q3FEL4 A0A182W8E1 A0A154PEP9 A0A182YQV0 A0A3L8DIT0 A0A026W5Y0 A0A151IFR4 K7IWF8 A0A232F1S2 A0A182NNN7 A0A1B0D2U3 A0A2J7RNU4 A0A0C9RN95 A0A151X567 A0A1J1I2L1 U5EZE5 A0A1L8DKA4 Q16VV3 A0A1S4FM88 A0A224XP98 A0A023EZ93 A0A182MN33 A0A1B6CTY0 A0A195BE85 A0A195FIU3 A0A0P6J545 A0A1L8DJX1 A0A158NR93 A0A182PVX8 A0A2J7Q8A1 A0A182GJV6 J3JXY8 N6TC26 U4UH84 E9IRY5 A0A151IU39 F4WB16 A0A1D2NCL7 A0A2P8YIK7 T1HQI4 A0A1D2NF22 A0A226EP64 A0A1B0CAI6 A0A2M4BPC2 E9FT77 A0A1B6DBG4 A0A2J7RNS7 A0A182FN08 A0A1Y1KQE4 A0A1D2MVX9 A0A0P5E628 B4LTL7 A0A0P5Y2T4 A0A0P6AEN3 A0A182WTW9 A0A182VL85 A0A182HUI5 Q7PNB3 B4JDP8 A0A067RK86
A0A2H1W793 A0A2W1B9R7 A0A194QMT9 A0A194QKR3 A0A212EWB0 A0A212EWD3 A0A0L7RE26 A0A1Y1ME42 A0A310SI06 A0A182K7G5 A0A067RMF2 A0A182TUQ6 A0A0M9A3Q8 A0A182HQX0 A0A182KJJ7 A0A182UZ84 A0A182J4G5 A0A182WXJ4 Q7PMH9 W5JRD6 A0A2M4BM64 A0A2A3EDW6 A0A2M4BLB4 A0A2M4BLB3 A0A2M4BLB8 A0A182QLD0 A0A023ET26 A0A087ZYQ7 E2A0F3 E2C7F5 A0A1W4WU99 A0A1W4WU80 B0W704 A0A1Q3FEL4 A0A182W8E1 A0A154PEP9 A0A182YQV0 A0A3L8DIT0 A0A026W5Y0 A0A151IFR4 K7IWF8 A0A232F1S2 A0A182NNN7 A0A1B0D2U3 A0A2J7RNU4 A0A0C9RN95 A0A151X567 A0A1J1I2L1 U5EZE5 A0A1L8DKA4 Q16VV3 A0A1S4FM88 A0A224XP98 A0A023EZ93 A0A182MN33 A0A1B6CTY0 A0A195BE85 A0A195FIU3 A0A0P6J545 A0A1L8DJX1 A0A158NR93 A0A182PVX8 A0A2J7Q8A1 A0A182GJV6 J3JXY8 N6TC26 U4UH84 E9IRY5 A0A151IU39 F4WB16 A0A1D2NCL7 A0A2P8YIK7 T1HQI4 A0A1D2NF22 A0A226EP64 A0A1B0CAI6 A0A2M4BPC2 E9FT77 A0A1B6DBG4 A0A2J7RNS7 A0A182FN08 A0A1Y1KQE4 A0A1D2MVX9 A0A0P5E628 B4LTL7 A0A0P5Y2T4 A0A0P6AEN3 A0A182WTW9 A0A182VL85 A0A182HUI5 Q7PNB3 B4JDP8 A0A067RK86
Pubmed
20053988
19121390
28756777
26354079
22118469
28004739
+ More
24845553 20966253 12364791 14747013 17210077 20920257 23761445 24945155 20798317 25244985 30249741 24508170 20075255 28648823 17510324 25474469 26999592 21347285 26483478 22516182 23537049 21282665 21719571 27289101 29403074 21292972 17994087 18057021
24845553 20966253 12364791 14747013 17210077 20920257 23761445 24945155 20798317 25244985 30249741 24508170 20075255 28648823 17510324 25474469 26999592 21347285 26483478 22516182 23537049 21282665 21719571 27289101 29403074 21292972 17994087 18057021
EMBL
AB515345
BAI66271.1
BABH01005172
BABH01005173
BABH01005174
BABH01005175
+ More
GDQN01005231 JAT85823.1 GDQN01006449 GDQN01002999 JAT84605.1 JAT88055.1 NWSH01000912 PCG73563.1 RSAL01000046 RVE50581.1 ODYU01006768 SOQ48917.1 KZ150380 PZC71095.1 KQ461194 KPJ06853.1 KQ458671 KPJ05505.1 AGBW02012020 OWR45788.1 OWR45789.1 KQ414613 KOC69079.1 GEZM01036247 JAV82860.1 KQ759870 OAD62418.1 KK852424 KDR24183.1 KQ435765 KOX75333.1 APCN01001607 AAAB01008980 EAA13815.3 ADMH02000643 ETN65470.1 GGFJ01004717 MBW53858.1 KZ288269 PBC29900.1 GGFJ01004718 MBW53859.1 GGFJ01004719 MBW53860.1 GGFJ01004716 MBW53857.1 AXCN02001996 GAPW01001160 JAC12438.1 GL435586 EFN73090.1 GL453369 EFN76142.1 DS231851 EDS37278.1 GFDL01009059 JAV25986.1 KQ434875 KZC09768.1 QOIP01000007 RLU20261.1 KK107390 EZA51413.1 KQ977771 KYN00005.1 NNAY01001229 OXU24681.1 AJVK01002848 AJVK01002849 NEVH01002148 PNF42494.1 GBYB01014957 JAG84724.1 KQ982540 KYQ55429.1 CVRI01000038 CRK93828.1 GANO01000941 JAB58930.1 GFDF01007314 JAV06770.1 CH477580 EAT38706.2 GFTR01006575 JAW09851.1 GBBI01004172 JAC14540.1 AXCM01005076 GEDC01020342 JAS16956.1 KQ976504 KYM82896.1 KQ981522 KYN40585.1 GDUN01000692 JAN95227.1 GFDF01007315 JAV06769.1 ADTU01023830 NEVH01016966 PNF24814.1 JXUM01068925 JXUM01068926 JXUM01068927 KQ562525 KXJ75697.1 BT128110 AEE63071.1 APGK01043534 APGK01043535 APGK01043536 KB741015 ENN75288.1 KB632161 ERL89265.1 GL765283 EFZ16682.1 KQ980970 KYN10930.1 GL888056 EGI68636.1 LJIJ01000091 ODN02981.1 PYGN01000568 PSN44095.1 ACPB03023768 LJIJ01000063 ODN03822.1 LNIX01000002 OXA59423.1 AJWK01003826 GGFJ01005650 MBW54791.1 GL732524 EFX89326.1 GEDC01014353 JAS22945.1 PNF42490.1 GEZM01076808 JAV63633.1 LJIJ01000462 ODM97219.1 GDIP01146046 JAJ77356.1 CH940649 EDW64990.1 KRF81970.1 KRF81971.1 GDIP01063756 LRGB01003123 JAM39959.1 KZS04220.1 GDIP01030748 JAM72967.1 APCN01002463 AAAB01008964 EAA12460.5 CH916368 EDW03418.1 KDR24217.1
GDQN01005231 JAT85823.1 GDQN01006449 GDQN01002999 JAT84605.1 JAT88055.1 NWSH01000912 PCG73563.1 RSAL01000046 RVE50581.1 ODYU01006768 SOQ48917.1 KZ150380 PZC71095.1 KQ461194 KPJ06853.1 KQ458671 KPJ05505.1 AGBW02012020 OWR45788.1 OWR45789.1 KQ414613 KOC69079.1 GEZM01036247 JAV82860.1 KQ759870 OAD62418.1 KK852424 KDR24183.1 KQ435765 KOX75333.1 APCN01001607 AAAB01008980 EAA13815.3 ADMH02000643 ETN65470.1 GGFJ01004717 MBW53858.1 KZ288269 PBC29900.1 GGFJ01004718 MBW53859.1 GGFJ01004719 MBW53860.1 GGFJ01004716 MBW53857.1 AXCN02001996 GAPW01001160 JAC12438.1 GL435586 EFN73090.1 GL453369 EFN76142.1 DS231851 EDS37278.1 GFDL01009059 JAV25986.1 KQ434875 KZC09768.1 QOIP01000007 RLU20261.1 KK107390 EZA51413.1 KQ977771 KYN00005.1 NNAY01001229 OXU24681.1 AJVK01002848 AJVK01002849 NEVH01002148 PNF42494.1 GBYB01014957 JAG84724.1 KQ982540 KYQ55429.1 CVRI01000038 CRK93828.1 GANO01000941 JAB58930.1 GFDF01007314 JAV06770.1 CH477580 EAT38706.2 GFTR01006575 JAW09851.1 GBBI01004172 JAC14540.1 AXCM01005076 GEDC01020342 JAS16956.1 KQ976504 KYM82896.1 KQ981522 KYN40585.1 GDUN01000692 JAN95227.1 GFDF01007315 JAV06769.1 ADTU01023830 NEVH01016966 PNF24814.1 JXUM01068925 JXUM01068926 JXUM01068927 KQ562525 KXJ75697.1 BT128110 AEE63071.1 APGK01043534 APGK01043535 APGK01043536 KB741015 ENN75288.1 KB632161 ERL89265.1 GL765283 EFZ16682.1 KQ980970 KYN10930.1 GL888056 EGI68636.1 LJIJ01000091 ODN02981.1 PYGN01000568 PSN44095.1 ACPB03023768 LJIJ01000063 ODN03822.1 LNIX01000002 OXA59423.1 AJWK01003826 GGFJ01005650 MBW54791.1 GL732524 EFX89326.1 GEDC01014353 JAS22945.1 PNF42490.1 GEZM01076808 JAV63633.1 LJIJ01000462 ODM97219.1 GDIP01146046 JAJ77356.1 CH940649 EDW64990.1 KRF81970.1 KRF81971.1 GDIP01063756 LRGB01003123 JAM39959.1 KZS04220.1 GDIP01030748 JAM72967.1 APCN01002463 AAAB01008964 EAA12460.5 CH916368 EDW03418.1 KDR24217.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000053825 UP000075881 UP000027135 UP000075902 UP000053105 UP000075840 UP000075882 UP000075903 UP000075880 UP000076407 UP000007062 UP000000673 UP000242457 UP000075886 UP000005203 UP000000311 UP000008237 UP000192223 UP000002320 UP000075920 UP000076502 UP000076408 UP000279307 UP000053097 UP000078542 UP000002358 UP000215335 UP000075884 UP000092462 UP000235965 UP000075809 UP000183832 UP000008820 UP000075883 UP000078540 UP000078541 UP000005205 UP000075885 UP000069940 UP000249989 UP000019118 UP000030742 UP000078492 UP000007755 UP000094527 UP000245037 UP000015103 UP000198287 UP000092461 UP000000305 UP000069272 UP000008792 UP000076858 UP000001070
UP000053825 UP000075881 UP000027135 UP000075902 UP000053105 UP000075840 UP000075882 UP000075903 UP000075880 UP000076407 UP000007062 UP000000673 UP000242457 UP000075886 UP000005203 UP000000311 UP000008237 UP000192223 UP000002320 UP000075920 UP000076502 UP000076408 UP000279307 UP000053097 UP000078542 UP000002358 UP000215335 UP000075884 UP000092462 UP000235965 UP000075809 UP000183832 UP000008820 UP000075883 UP000078540 UP000078541 UP000005205 UP000075885 UP000069940 UP000249989 UP000019118 UP000030742 UP000078492 UP000007755 UP000094527 UP000245037 UP000015103 UP000198287 UP000092461 UP000000305 UP000069272 UP000008792 UP000076858 UP000001070
Interpro
Gene 3D
ProteinModelPortal
D2KXB2
H9JLS4
A0A1E1WFQ5
A0A1E1WCD1
A0A2A4JQ46
A0A3S2P2C8
+ More
A0A2H1W793 A0A2W1B9R7 A0A194QMT9 A0A194QKR3 A0A212EWB0 A0A212EWD3 A0A0L7RE26 A0A1Y1ME42 A0A310SI06 A0A182K7G5 A0A067RMF2 A0A182TUQ6 A0A0M9A3Q8 A0A182HQX0 A0A182KJJ7 A0A182UZ84 A0A182J4G5 A0A182WXJ4 Q7PMH9 W5JRD6 A0A2M4BM64 A0A2A3EDW6 A0A2M4BLB4 A0A2M4BLB3 A0A2M4BLB8 A0A182QLD0 A0A023ET26 A0A087ZYQ7 E2A0F3 E2C7F5 A0A1W4WU99 A0A1W4WU80 B0W704 A0A1Q3FEL4 A0A182W8E1 A0A154PEP9 A0A182YQV0 A0A3L8DIT0 A0A026W5Y0 A0A151IFR4 K7IWF8 A0A232F1S2 A0A182NNN7 A0A1B0D2U3 A0A2J7RNU4 A0A0C9RN95 A0A151X567 A0A1J1I2L1 U5EZE5 A0A1L8DKA4 Q16VV3 A0A1S4FM88 A0A224XP98 A0A023EZ93 A0A182MN33 A0A1B6CTY0 A0A195BE85 A0A195FIU3 A0A0P6J545 A0A1L8DJX1 A0A158NR93 A0A182PVX8 A0A2J7Q8A1 A0A182GJV6 J3JXY8 N6TC26 U4UH84 E9IRY5 A0A151IU39 F4WB16 A0A1D2NCL7 A0A2P8YIK7 T1HQI4 A0A1D2NF22 A0A226EP64 A0A1B0CAI6 A0A2M4BPC2 E9FT77 A0A1B6DBG4 A0A2J7RNS7 A0A182FN08 A0A1Y1KQE4 A0A1D2MVX9 A0A0P5E628 B4LTL7 A0A0P5Y2T4 A0A0P6AEN3 A0A182WTW9 A0A182VL85 A0A182HUI5 Q7PNB3 B4JDP8 A0A067RK86
A0A2H1W793 A0A2W1B9R7 A0A194QMT9 A0A194QKR3 A0A212EWB0 A0A212EWD3 A0A0L7RE26 A0A1Y1ME42 A0A310SI06 A0A182K7G5 A0A067RMF2 A0A182TUQ6 A0A0M9A3Q8 A0A182HQX0 A0A182KJJ7 A0A182UZ84 A0A182J4G5 A0A182WXJ4 Q7PMH9 W5JRD6 A0A2M4BM64 A0A2A3EDW6 A0A2M4BLB4 A0A2M4BLB3 A0A2M4BLB8 A0A182QLD0 A0A023ET26 A0A087ZYQ7 E2A0F3 E2C7F5 A0A1W4WU99 A0A1W4WU80 B0W704 A0A1Q3FEL4 A0A182W8E1 A0A154PEP9 A0A182YQV0 A0A3L8DIT0 A0A026W5Y0 A0A151IFR4 K7IWF8 A0A232F1S2 A0A182NNN7 A0A1B0D2U3 A0A2J7RNU4 A0A0C9RN95 A0A151X567 A0A1J1I2L1 U5EZE5 A0A1L8DKA4 Q16VV3 A0A1S4FM88 A0A224XP98 A0A023EZ93 A0A182MN33 A0A1B6CTY0 A0A195BE85 A0A195FIU3 A0A0P6J545 A0A1L8DJX1 A0A158NR93 A0A182PVX8 A0A2J7Q8A1 A0A182GJV6 J3JXY8 N6TC26 U4UH84 E9IRY5 A0A151IU39 F4WB16 A0A1D2NCL7 A0A2P8YIK7 T1HQI4 A0A1D2NF22 A0A226EP64 A0A1B0CAI6 A0A2M4BPC2 E9FT77 A0A1B6DBG4 A0A2J7RNS7 A0A182FN08 A0A1Y1KQE4 A0A1D2MVX9 A0A0P5E628 B4LTL7 A0A0P5Y2T4 A0A0P6AEN3 A0A182WTW9 A0A182VL85 A0A182HUI5 Q7PNB3 B4JDP8 A0A067RK86
PDB
6I2K
E-value=2.8983e-40,
Score=416
Ontologies
KEGG
GO
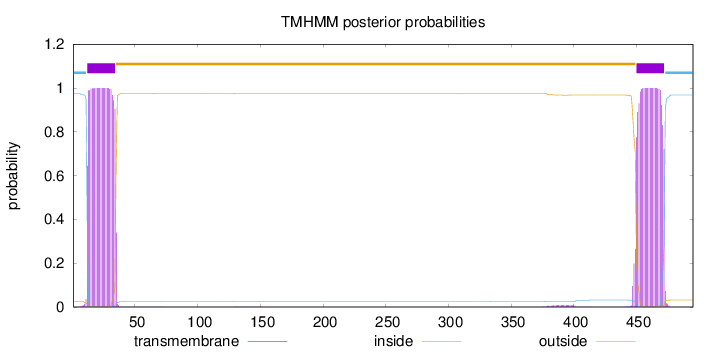
Topology
Length:
495
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.79481
Exp number, first 60 AAs:
22.86836
Total prob of N-in:
0.97536
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 449
TMhelix
450 - 472
inside
473 - 495
Population Genetic Test Statistics
Pi
228.420521
Theta
168.364418
Tajima's D
-2.334027
CLR
0.866595
CSRT
0.00244987750612469
Interpretation
Possibly Positive selection
